Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T33584
(Former ID: TTDC00250)
|
|||||
| Target Name |
Glutamate receptor AMPA 1 (GRIA1)
|
|||||
| Synonyms |
Glutamate receptor ionotropic, AMPA 1; Glutamate receptor 1; GluR-K1; GluR-A; GluR-1; GluA1; GLUR1; GLUH1; AMPA-selective glutamate receptor 1
Click to Show/Hide
|
|||||
| Gene Name |
GRIA1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Neuropathy [ICD-11: 8C0Z] | |||||
| Function |
L-glutamate acts as an excitatory neurotransmitter at many synapses in the central nervous system. Binding of the excitatory neurotransmitter L-glutamate induces a conformation change, leading to the opening of the cation channel, and thereby converts the chemical signal to an electrical impulse. The receptor then desensitizes rapidly and enters a transient inactive state, characterized by the presence of bound agonist. In the presence of CACNG4 or CACNG7 or CACNG8, shows resensitization which is characterized by a delayed accumulation of current flux upon continued application of glutamate. Ionotropic glutamate receptor.
Click to Show/Hide
|
|||||
| BioChemical Class |
Glutamate-gated ion channel
|
|||||
| UniProt ID | ||||||
| Sequence |
MQHIFAFFCTGFLGAVVGANFPNNIQIGGLFPNQQSQEHAAFRFALSQLTEPPKLLPQID
IVNISDSFEMTYRFCSQFSKGVYAIFGFYERRTVNMLTSFCGALHVCFITPSFPVDTSNQ FVLQLRPELQDALISIIDHYKWQKFVYIYDADRGLSVLQKVLDTAAEKNWQVTAVNILTT TEEGYRMLFQDLEKKKERLVVVDCESERLNAILGQIIKLEKNGIGYHYILANLGFMDIDL NKFKESGANVTGFQLVNYTDTIPAKIMQQWKNSDARDHTRVDWKRPKYTSALTYDGVKVM AEAFQSLRRQRIDISRRGNAGDCLANPAVPWGQGIDIQRALQQVRFEGLTGNVQFNEKGR RTNYTLHVIEMKHDGIRKIGYWNEDDKFVPAATDAQAGGDNSSVQNRTYIVTTILEDPYV MLKKNANQFEGNDRYEGYCVELAAEIAKHVGYSYRLEIVSDGKYGARDPDTKAWNGMVGE LVYGRADVAVAPLTITLVREEVIDFSKPFMSLGISIMIKKPQKSKPGVFSFLDPLAYEIW MCIVFAYIGVSVVLFLVSRFSPYEWHSEEFEEGRDQTTSDQSNEFGIFNSLWFSLGAFMQ QGCDISPRSLSGRIVGGVWWFFTLIIISSYTANLAAFLTVERMVSPIESAEDLAKQTEIA YGTLEAGSTKEFFRRSKIAVFEKMWTYMKSAEPSVFVRTTEEGMIRVRKSKGKYAYLLES TMNEYIEQRKPCDTMKVGGNLDSKGYGIATPKGSALRNPVNLAVLKLNEQGLLDKLKNKW WYDKGECGSGGGDSKDKTSALSLSNVAGVFYILIGGLGLAMLVALIEFCYKSRSESKRMK GFCLIPQQSINEAIRTSTLPRNSGAGASSGGSGENGRVVSHDFPKSMQSIPCMSHSSGMP LGATGL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T51WML | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | NBQX | Drug Info | Phase 1 | Neurological disorder | [4] | |
| Discontinued Drug(s) | [+] 3 Discontinued Drugs | + | ||||
| 1 | YM-90K | Drug Info | Discontinued in Phase 2 | Convulsion | [5] | |
| 2 | GYKI-52466 | Drug Info | Terminated | Alzheimer disease | [7], [8] | |
| 3 | SORETOLIDE | Drug Info | Terminated | Convulsion | [9] | |
| Mode of Action | [+] 5 Modes of Action | + | ||||
| Inhibitor | [+] 14 Inhibitor drugs | + | ||||
| 1 | NBQX | Drug Info | [10] | |||
| 2 | YM-90K | Drug Info | [11] | |||
| 3 | GYKI-52466 | Drug Info | [12] | |||
| 4 | GYKI-53655 | Drug Info | [13] | |||
| 5 | (S)-AMPA | Drug Info | [16] | |||
| 6 | (S)-WILLARDIINE | Drug Info | [17] | |||
| 7 | 7-chloro-3-hydroxyquinazoline-2,4-dione | Drug Info | [10] | |||
| 8 | Argiotoxin-636 | Drug Info | [18] | |||
| 9 | DNQX | Drug Info | [19] | |||
| 10 | N-(4-hydroxyphenylpropanyl)-spermine | Drug Info | [18] | |||
| 11 | Philanthotoxin-343 | Drug Info | [18] | |||
| 12 | Piriqualone | Drug Info | [20] | |||
| 13 | RPR-118723 | Drug Info | [21] | |||
| 14 | [3H]kainate | Drug Info | [23] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | SORETOLIDE | Drug Info | [14] | |||
| Agonist | [+] 2 Agonist drugs | + | ||||
| 1 | (S)-5-fluorowillardiine | Drug Info | [15] | |||
| 2 | [3H]AMPA | Drug Info | [15] | |||
| Antagonist | [+] 2 Antagonist drugs | + | ||||
| 1 | ATPO | Drug Info | [15] | |||
| 2 | [3H]CNQX | Drug Info | [15], [22] | |||
| Blocker (channel blocker) | [+] 1 Blocker (channel blocker) drugs | + | ||||
| 1 | joro toxin | Drug Info | [15] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cAMP signaling pathway | hsa04024 | Affiliated Target |
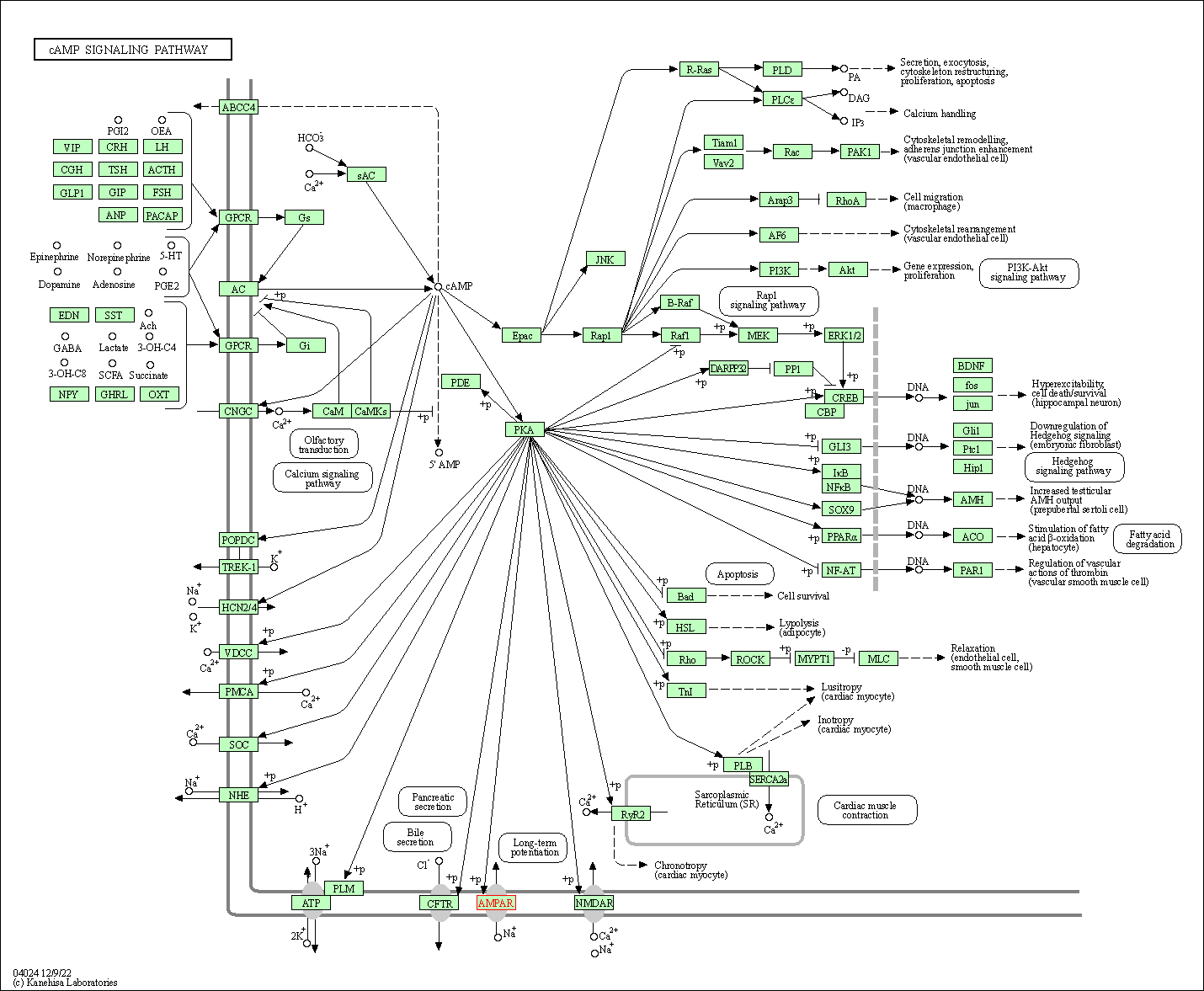
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Neuroactive ligand-receptor interaction | hsa04080 | Affiliated Target |
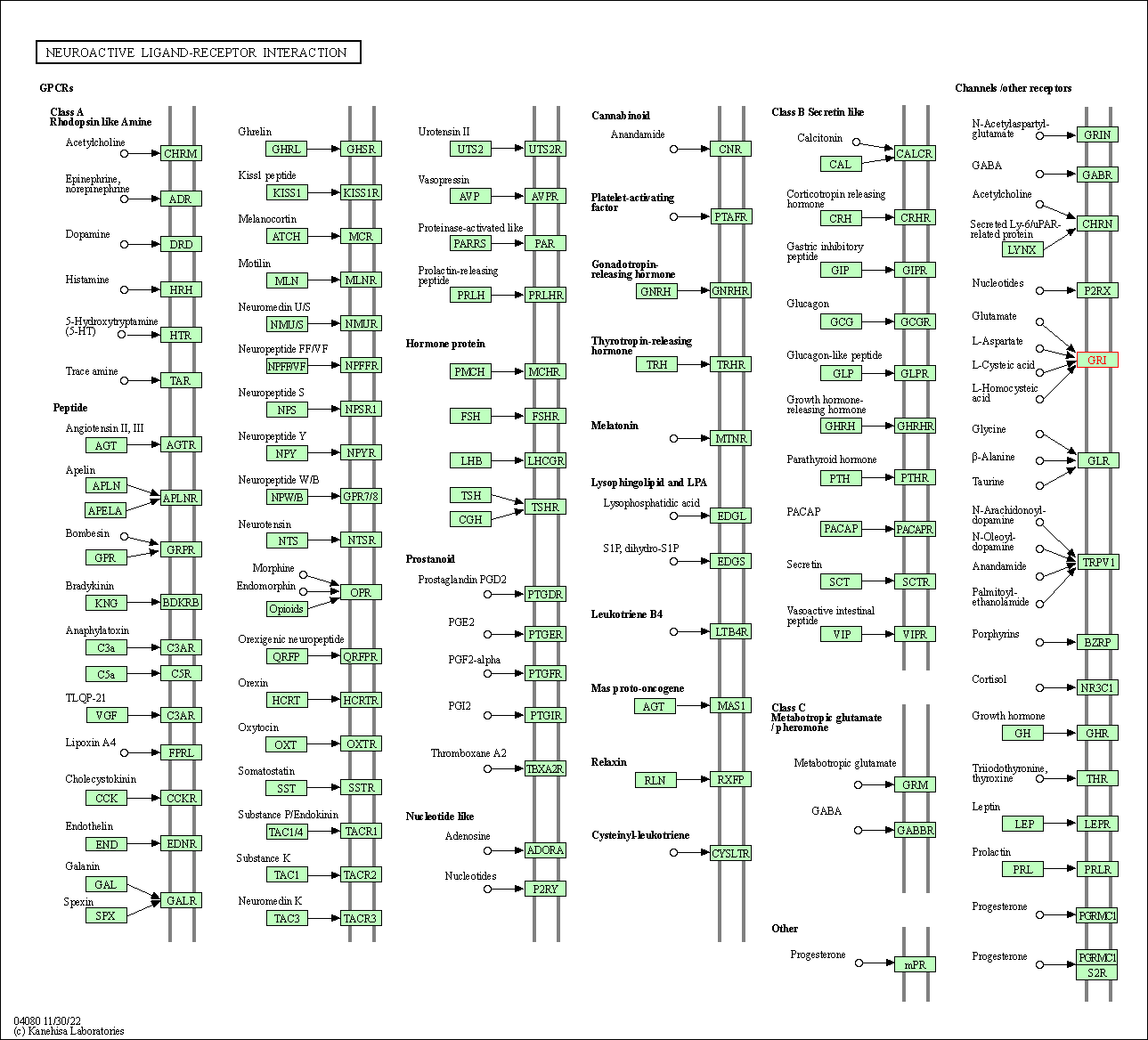
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Circadian entrainment | hsa04713 | Affiliated Target |
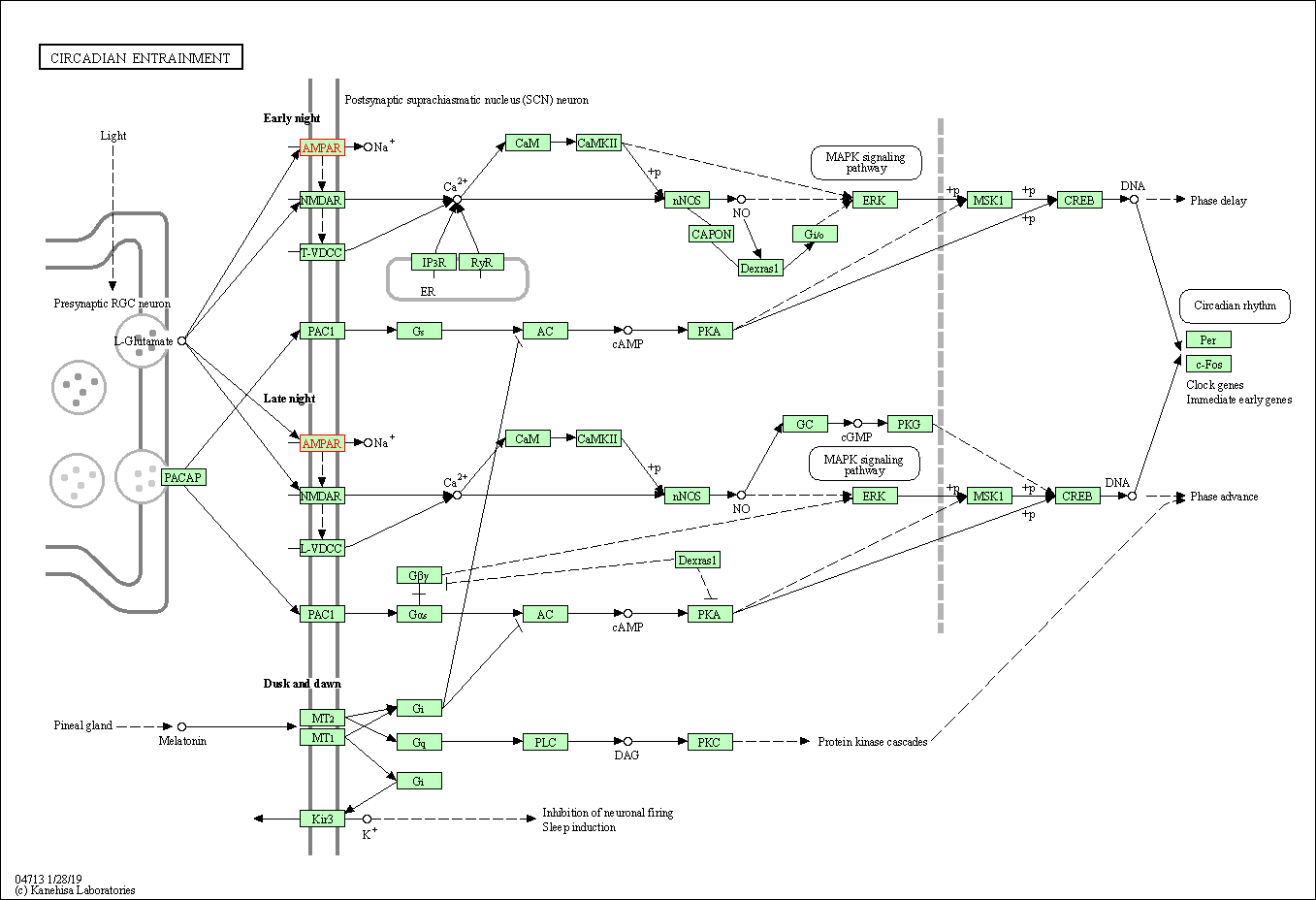
|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |
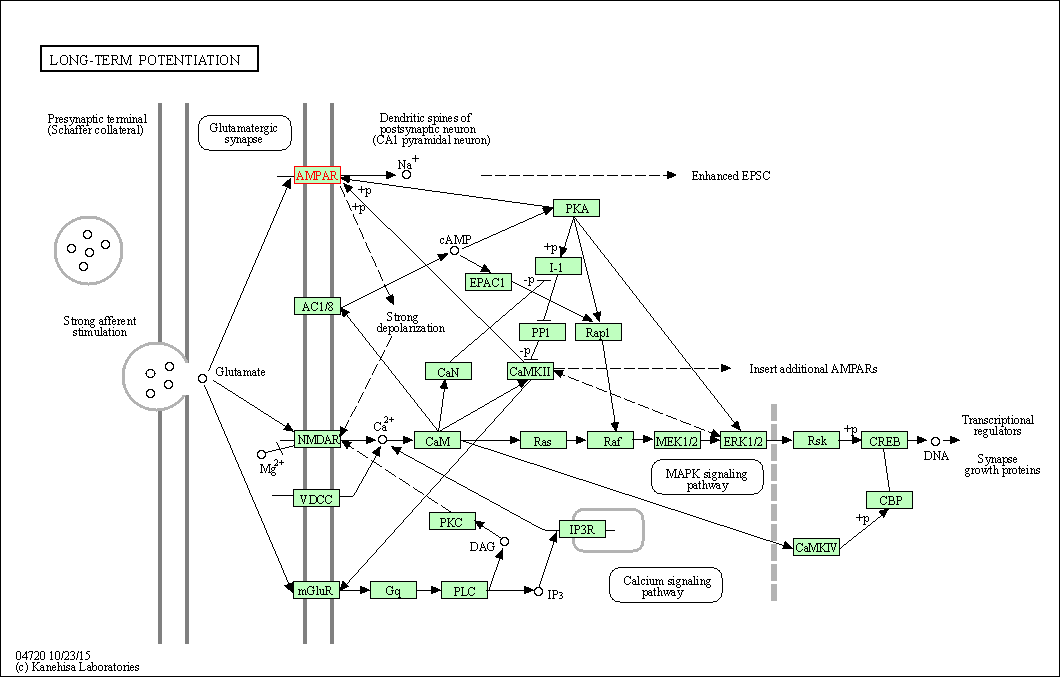
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Retrograde endocannabinoid signaling | hsa04723 | Affiliated Target |
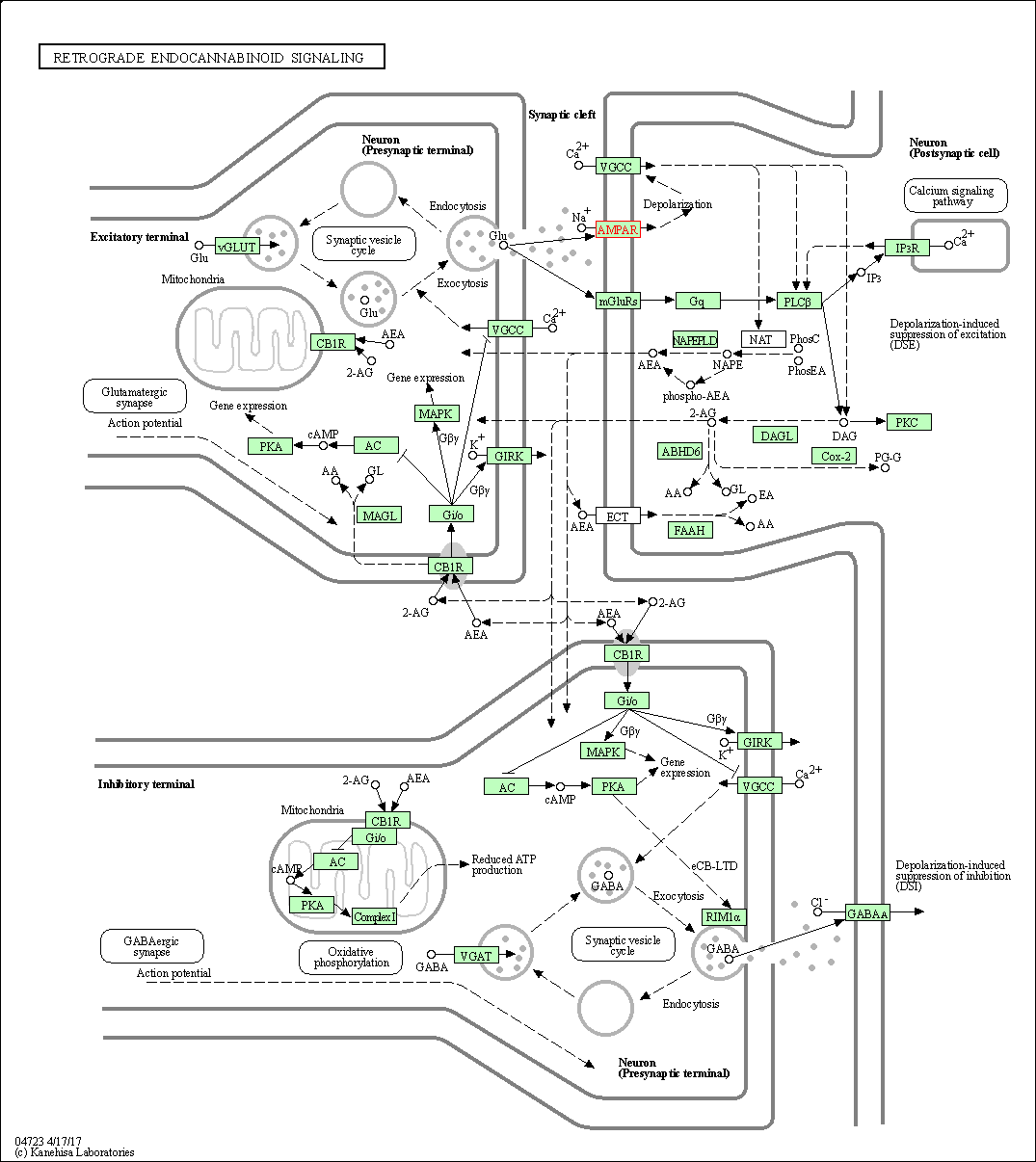
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Glutamatergic synapse | hsa04724 | Affiliated Target |
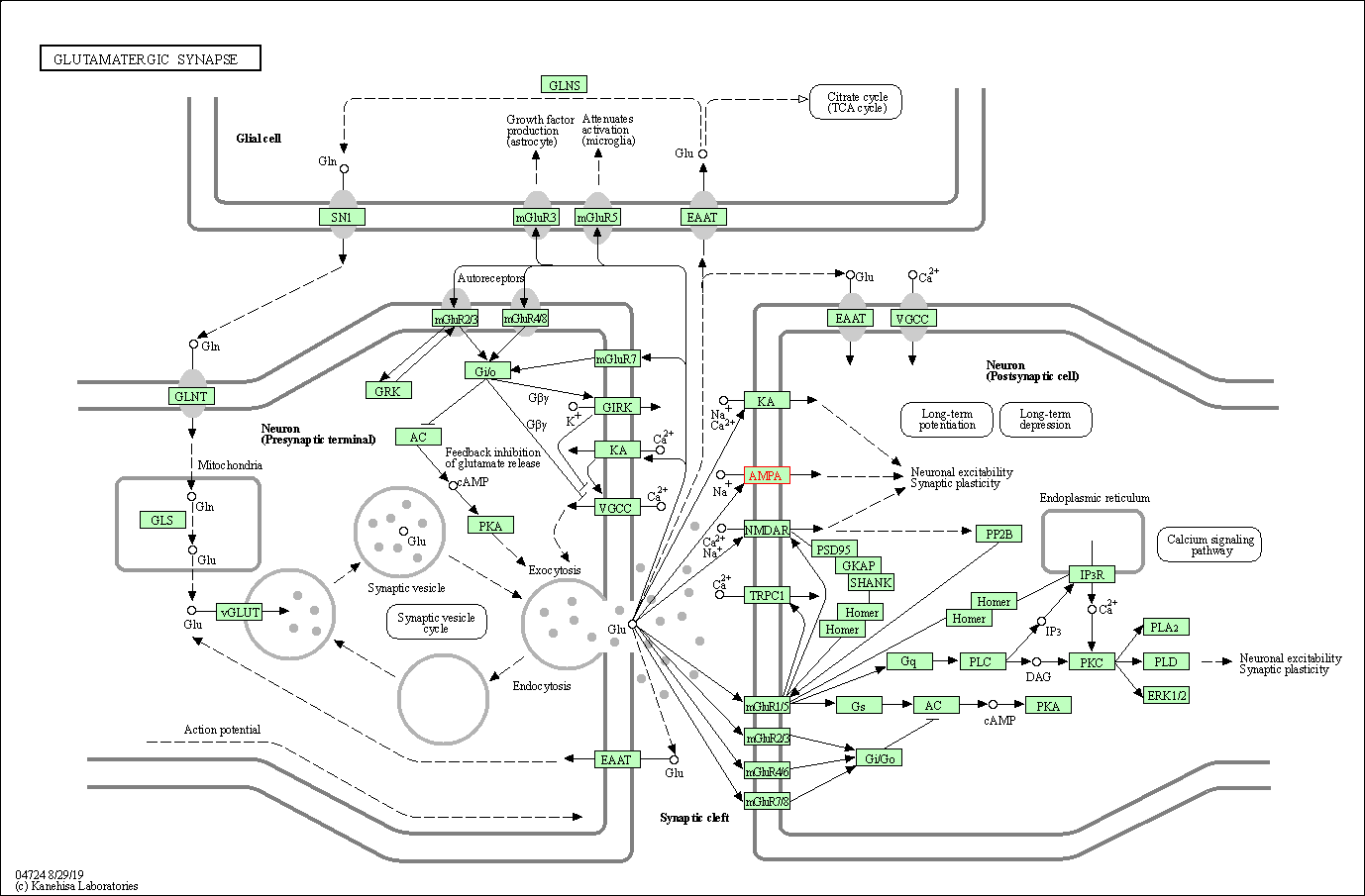
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
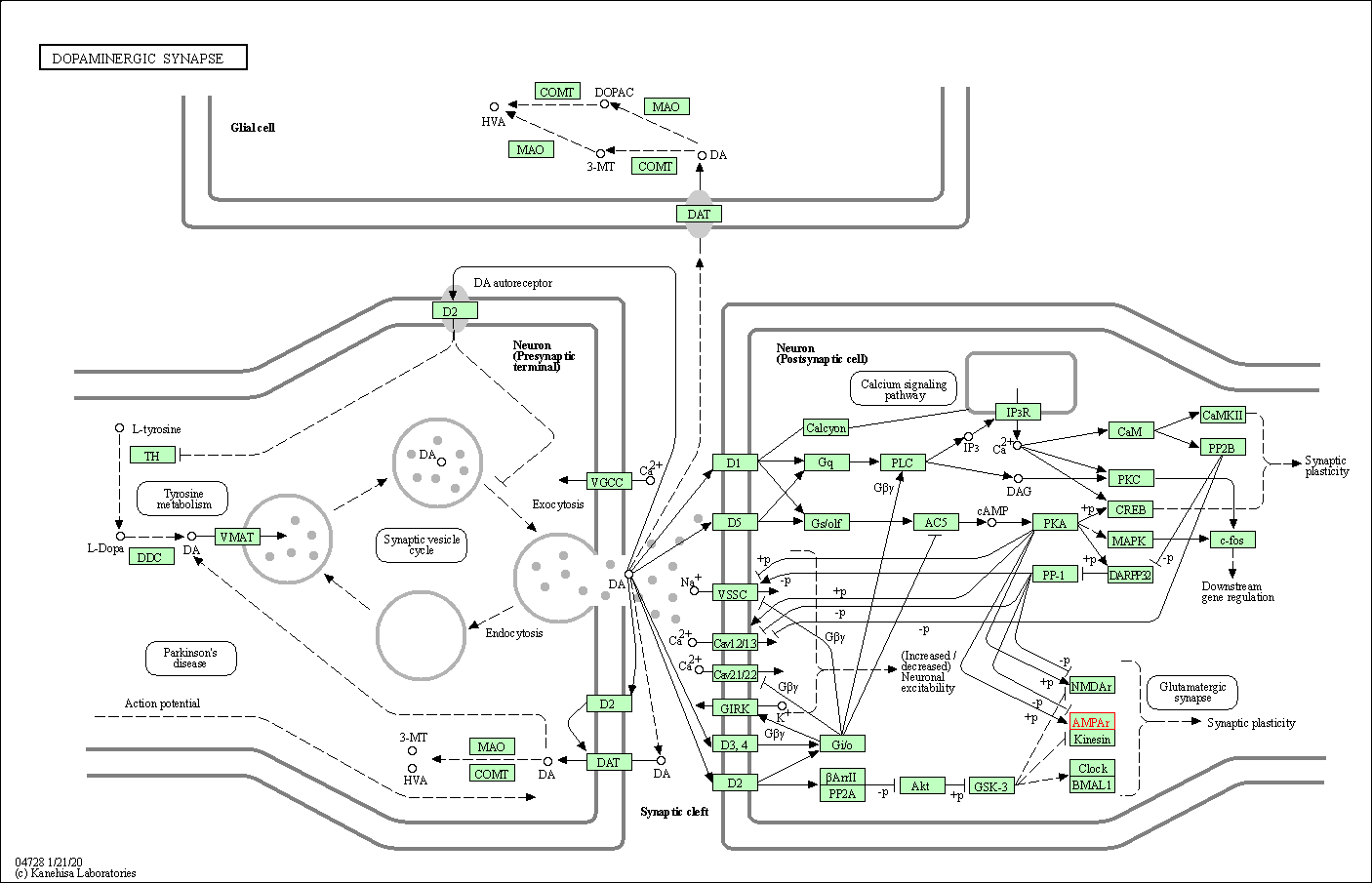
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Long-term depression | hsa04730 | Affiliated Target |
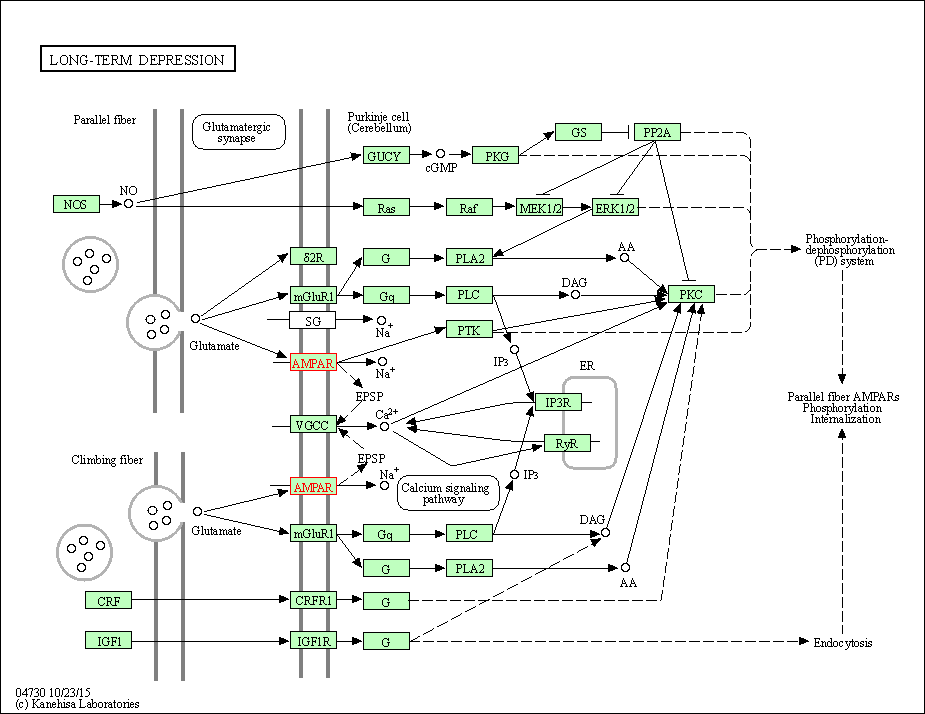
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 19 | Degree centrality | 2.04E-03 | Betweenness centrality | 5.37E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.16E-01 | Radiality | 1.38E+01 | Clustering coefficient | 2.28E-01 |
| Neighborhood connectivity | 1.42E+01 | Topological coefficient | 9.25E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Antibodies and venom peptides: new modalities for ion channels. Nat Rev Drug Discov. 2019 May;18(5):339-357. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7050). | |||||
| REF 3 | Nat Rev Drug Discov. 2013 Feb;12(2):87-90. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4264). | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002155) | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800012259) | |||||
| REF 7 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4210). | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800005470) | |||||
| REF 9 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002789) | |||||
| REF 10 | Structural investigation of the 7-chloro-3-hydroxy-1H-quinazoline-2,4-dione scaffold to obtain AMPA and kainate receptor selective antagonists. Syn... J Med Chem. 2006 Oct 5;49(20):6015-26. | |||||
| REF 11 | Synthesis and AMPA receptor antagonistic activity of a novel 7-imidazolyl-6-trifluoromethyl quinoxalinecarboxylic acid with a substituted phenyl gr... Bioorg Med Chem Lett. 2004 Oct 18;14(20):5107-11. | |||||
| REF 12 | New 7,8-ethylenedioxy-2,3-benzodiazepines as noncompetitive AMPA receptor antagonists. Bioorg Med Chem Lett. 2006 Jan 1;16(1):167-70. | |||||
| REF 13 | Substituted 1,2-dihydrophthalazines: potent, selective, and noncompetitive inhibitors of the AMPA receptor. J Med Chem. 1996 Jan 19;39(2):343-6. | |||||
| REF 14 | Preclinical pharmacology of perampanel, a selective non-competitive AMPA receptor antagonist. Acta Neurol Scand Suppl. 2013;(197):19-24. | |||||
| REF 15 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 444). | |||||
| REF 16 | Synthesis and pharmacology of willardiine derivatives acting as antagonists of kainate receptors. J Med Chem. 2005 Dec 1;48(24):7867-81. | |||||
| REF 17 | Synthesis of willardiine and 6-azawillardiine analogs: pharmacological characterization on cloned homomeric human AMPA and kainate receptor subtypes. J Med Chem. 1997 Oct 24;40(22):3645-50. | |||||
| REF 18 | Developing a complete pharmacology for AMPA receptors: a perspective on subtype-selective ligands. Bioorg Med Chem. 2010 Feb 15;18(4):1381-7. | |||||
| REF 19 | Synthesis of chiral 1-(2'-amino-2'-carboxyethyl)-1,4-dihydro-6,7-quinoxaline-2,3-diones: alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate recep... J Med Chem. 1996 Oct 25;39(22):4430-8. | |||||
| REF 20 | Atropisomeric quinazolin-4-one derivatives are potent noncompetitive alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor antag... Bioorg Med Chem Lett. 2001 Jan 22;11(2):177-81. | |||||
| REF 21 | Indeno[1,2-b]pyrazin-2,3-diones: a new class of antagonists at the glycine site of the NMDA receptor with potent in vivo activity. J Med Chem. 2000 Jun 15;43(12):2371-81. | |||||
| REF 22 | Structure-activity relationship studies on N3-substituted willardiine derivatives acting as AMPA or kainate receptor antagonists. J Med Chem. 2006 Apr 20;49(8):2579-92. | |||||
| REF 23 | 1H-cyclopentapyrimidine-2,4(1H,3H)-dione-related ionotropic glutamate receptors ligands. structure-activity relationships and identification of pot... J Med Chem. 2008 Oct 23;51(20):6614-8. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

