| Drug General Information |
| Drug ID |
D0S5RC
|
| Former ID |
DCL000007
|
| Drug Name |
SNS-032
|
| Synonyms |
BMS-387032; BMS-387072; BMS-387032, SNS-032; N-[5-[[[5-(1,1-Dimethylethyl)-2-oxazolyl]methyl]thio]-2-thiazolyl]-4-piperidinecarboxamide L-tartaric acid salt (2:1); N-(5-(((5-(1,1-Dimethylethyl)-2-oxazolyl)methyl)thio)-2-thiazolyl)-4-piperidinecarboxamide; N-[5-[(5-tert-butyl-1,3-oxazol-2-yl)methylsulfanyl]-1,3-thiazol-2-yl]piperidine-4-carboxamide; (2R,3R)-2,3-dihydroxybutanedioic acid
|
| Drug Type |
Small molecular drug
|
| Therapeutic Class |
Anticancer Agents
|
| Company |
Sunesis; Bristol Myers Squibb
|
| Structure |
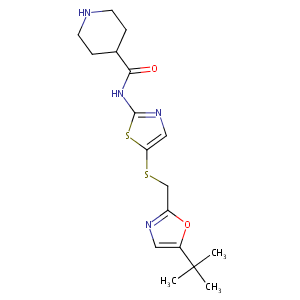
|
Download
2D MOL
3D MOL
|
| Formula |
C17H24N4O2S2
|
| InChI |
InChI=1S/C17H24N4O2S2/c1-17(2,3)12-8-19-13(23-12)10-24-14-9-20-16(25-14)21-15(22)11-4-6-18-7-5-11/h8-9,11,18H,4-7,10H2,1-3H3,(H,20,21,22)
|
| InChIKey |
OUSFTKFNBAZUKL-UHFFFAOYSA-N
|
| PubChem Compound ID |
|
| PubChem Substance ID |
8035540, 10043749, 14755895, 36070876, 50069793, 50100091, 50550338, 57352548, 93309880, 99437001, 99451015, 103229612, 103853138, 103905658, 111614343, 124757032, 124772085, 125163836, 125571016, 126577741, 126666806, 126729171, 126735498, 131480704, 135215937, 136340213, 136367332, 136367781, 137276011, 142439008, 143497596, 144233365, 152038136, 152240014, 152258053, 152344090, 160646891, 162011577, 162037466, 162202565, 162840915, 163412142, 164765032, 165237971, 174007077, 174529000, 177748740, 178102298, 180386839, 184819914
|
| Target and Pathway |
| References |