| Drug General Information |
| Drug ID |
D01HNP
|
| Former ID |
DNC003435
|
| Drug Name |
Hydroxyacetic Acid
|
| Drug Type |
Small molecular drug
|
| Structure |
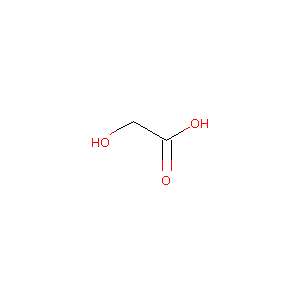
|
Download
2D MOL
3D MOL
|
| Formula |
C2H4O3
|
| Canonical SMILES |
C(C(=O)O)O
|
| InChI |
1S/C2H4O3/c3-1-2(4)5/h3H,1H2,(H,4,5)
|
| InChIKey |
AEMRFAOFKBGASW-UHFFFAOYSA-N
|
| CAS Number |
CAS 79-14-1
|
| PubChem Compound ID |
|
| PubChem Substance ID |
3460, 6350, 67104, 589599, 623904, 827351, 841751, 3135611, 4523739, 7887947, 8028465, 8137761, 8145512, 8150775, 10536092, 15119621, 24439119, 24847624, 24866341, 24866342, 24869373, 24870413, 24895337, 24898199, 26710336, 26713486, 26717671, 29216298, 29216299, 37336365, 37356959, 37460647, 46508186, 48416874, 48420440, 48423471, 49856273, 51074820, 56355025, 57320457, 57650858, 58106660, 75982826, 81058771, 85086452, 85086453, 85165053, 85244535, 85261655, 87325945
|
| Target and Pathway |
| References |