Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T00895
(Former ID: TTDC00066)
|
|||||
| Target Name |
Protein kinase C epsilon (PRKCE)
|
|||||
| Synonyms |
Protein kinase C epsilon type; PKCE; PKC epsilon; NPKC-epsilon
Click to Show/Hide
|
|||||
| Gene Name |
PRKCE
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Anxiety disorder [ICD-11: 6B00-6B0Z] | |||||
| 2 | Malaria [ICD-11: 1F40-1F45] | |||||
| Function |
Mediates cell adhesion to the extracellular matrix via integrin-dependent signaling, by mediating angiotensin-2-induced activation of integrin beta-1 (ITGB1) in cardiac fibroblasts. Phosphorylates MARCKS, which phosphorylates and activates PTK2/FAK, leading to the spread of cardiomyocytes. Involved in the control of the directional transport of ITGB1 in mesenchymal cells by phosphorylating vimentin (VIM), an intermediate filament (IF) protein. In epithelial cells, associates with and phosphorylates keratin-8 (KRT8), which induces targeting of desmoplakin at desmosomes and regulates cell-cell contact. Phosphorylates IQGAP1, which binds to CDC42, mediating epithelial cell-cell detachment prior to migration. In HeLa cells, contributes to hepatocyte growth factor (HGF)-induced cell migration, and in human corneal epithelial cells, plays a critical role in wound healing after activation by HGF. During cytokinesis, forms a complex with YWHAB, which is crucial for daughter cell separation, and facilitates abscission by a mechanism which may implicate the regulation of RHOA. In cardiac myocytes, regulates myofilament function and excitation coupling at the Z-lines, where it is indirectly associated with F-actin via interaction with COPB1. During endothelin-induced cardiomyocyte hypertrophy, mediates activation of PTK2/FAK, which is critical for cardiomyocyte survival and regulation of sarcomere length. Plays a role in the pathogenesis of dilated cardiomyopathy via persistent phosphorylation of troponin I (TNNI3). Involved in nerve growth factor (NFG)-induced neurite outgrowth and neuron morphological change independently of its kinase activity, by inhibition of RHOA pathway, activation of CDC42 and cytoskeletal rearrangement. May be involved in presynaptic facilitation by mediating phorbol ester-induced synaptic potentiation. Phosphorylates gamma-aminobutyric acid receptor subunit gamma-2 (GABRG2), which reduces the response of GABA receptors to ethanol and benzodiazepines and may mediate acute tolerance to the intoxicating effects of ethanol. Upon PMA treatment, phosphorylates the capsaicin- and heat-activated cation channel TRPV1, which is required for bradykinin-induced sensitization of the heat response in nociceptive neurons. Is able to form a complex with PDLIM5 and N-type calcium channel, and may enhance channel activities and potentiates fast synaptic transmission by phosphorylating the pore-forming alpha subunit CACNA1B (CaV2. 2). In prostate cancer cells, interacts with and phosphorylates STAT3, which increases DNA-binding and transcriptional activity of STAT3 and seems to be essential for prostate cancer cell invasion. Downstream of TLR4, plays an important role in the lipopolysaccharide (LPS)-induced immune response by phosphorylating and activating TICAM2/TRAM, which in turn activates the transcription factor IRF3 and subsequent cytokines production. In differentiating erythroid progenitors, is regulated by EPO and controls the protection against the TNFSF10/TRAIL-mediated apoptosis, via BCL2. May be involved in the regulation of the insulin-induced phosphorylation and activation of AKT1. Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays essential roles in the regulation of multiple cellular processes linked to cytoskeletal proteins, such as cell adhesion, motility, migration and cell cycle, functions in neuron growth and ion channel regulation, and is involved in immune response, cancer cell invasion and regulation of apoptosis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.13
|
|||||
| Sequence |
MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKT
NSPAWHDEFVTDVCNGRKIELAVFHDAPIGYDDFVANCTIQFEELLQNGSRHFEDWIDLE PEGRVYVIIDLSGSSGEAPKDNEERVFRERMRPRKRQGAVRRRVHQVNGHKFMATYLRQP TYCSHCRDFIWGVIGKQGYQCQVCTCVVHKRCHELIITKCAGLKKQETPDQVGSQRFSVN MPHKFGIHNYKVPTFCDHCGSLLWGLLRQGLQCKVCKMNVHRRCETNVAPNCGVDARGIA KVLADLGVTPDKITNSGQRRKKLIAGAESPQPASGSSPSEEDRSKSAPTSPCDQEIKELE NNIRKALSFDNRGEEHRAASSPDGQLMSPGENGEVRQGQAKRLGLDEFNFIKVLGKGSFG KVMLAELKGKDEVYAVKVLKKDVILQDDDVDCTMTEKRILALARKHPYLTQLYCCFQTKD RLFFVMEYVNGGDLMFQIQRSRKFDEPRSRFYAAEVTSALMFLHQHGVIYRDLKLDNILL DAEGHCKLADFGMCKEGILNGVTTTTFCGTPDYIAPEILQELEYGPSVDWWALGVLMYEM MAGQPPFEADNEDDLFESILHDDVLYPVWLSKEAVSILKAFMTKNPHKRLGCVASQNGED AIKQHPFFKEIDWVLLEQKKIKPPFKPRIKTKRDVNNFDQDFTREEPVLTLVDEAIVKQI NQEEFKGFSYFGEDLMP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T62Y4H | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Bryostatin-1 | Drug Info | Phase 2 | Alzheimer disease | [4], [5] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | CDE-6960 | Drug Info | Terminated | Inflammation | [6] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Activator | [+] 2 Activator drugs | + | ||||
| 1 | Bryostatin-1 | Drug Info | [7] | |||
| 2 | Phorbol 12-myristate | Drug Info | [7] | |||
| Inhibitor | [+] 19 Inhibitor drugs | + | ||||
| 1 | CDE-6960 | Drug Info | [8] | |||
| 2 | RO-320432 | Drug Info | [9] | |||
| 3 | 2,3,3-Triphenyl-acrylonitrile | Drug Info | [10] | |||
| 4 | 2-(4-Hydroxy-phenyl)-3,3-diphenyl-acrylonitrile | Drug Info | [10] | |||
| 5 | 3,3-Bis-(4-hydroxy-phenyl)-2-phenyl-acrylonitrile | Drug Info | [10] | |||
| 6 | 3,3-Bis-(4-methoxy-phenyl)-2-phenyl-acrylonitrile | Drug Info | [10] | |||
| 7 | 3,4-di-(4-methoxyphenyl)-1H-pyrrole-2,5-dione | Drug Info | [11] | |||
| 8 | 3,4-diphenyl-1H-pyrrole-2,5-dione | Drug Info | [11] | |||
| 9 | 3-(4-Hydroxy-phenyl)-2,3-diphenyl-acrylonitrile | Drug Info | [10] | |||
| 10 | 3-(4-methoxyphenyl)-4-phenyl-1H-pyrrole-2,5-dione | Drug Info | [11] | |||
| 11 | 3-(indole-3-yl)-4-phenyl-1H-pyrrole-2,5-dione | Drug Info | [11] | |||
| 12 | 4-cycloheptyliden(4-hydroxyphenyl)methylphenol | Drug Info | [10] | |||
| 13 | 4-[1-(4-hydroxyphenyl)-3-methyl-1-butenyl]phenol | Drug Info | [10] | |||
| 14 | LY-326449 | Drug Info | [12] | |||
| 15 | RO-316233 | Drug Info | [13] | |||
| 16 | Ro-32-0557 | Drug Info | [9] | |||
| 17 | [2,2':5',2'']Terthiophen-4-yl-methanol | Drug Info | [14] | |||
| 18 | [2,2':5',2'']Terthiophene-4,5''-dicarbaldehyde | Drug Info | [14] | |||
| 19 | [2,2':5',2'']Terthiophene-4-carbaldehyde | Drug Info | [14] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: adenosine diphosphate | Ligand Info | |||||
| Structure Description | Structure of a peptide-substrate bound to PKCiota core kinase domain | PDB:5LIH | ||||
| Method | X-ray diffraction | Resolution | 3.25 Å | Mutation | No | [15] |
| PDB Sequence |
RMRPFKRQGS
11 VRR
|
|||||
|
|
||||||
| Ligand Name: Phosphonothreonine | Ligand Info | |||||
| Structure Description | Structure of a peptide-substrate bound to PKCiota core kinase domain | PDB:5LIH | ||||
| Method | X-ray diffraction | Resolution | 3.25 Å | Mutation | No | [15] |
| PDB Sequence |
RMRPFKRQGS
11 VRR
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |
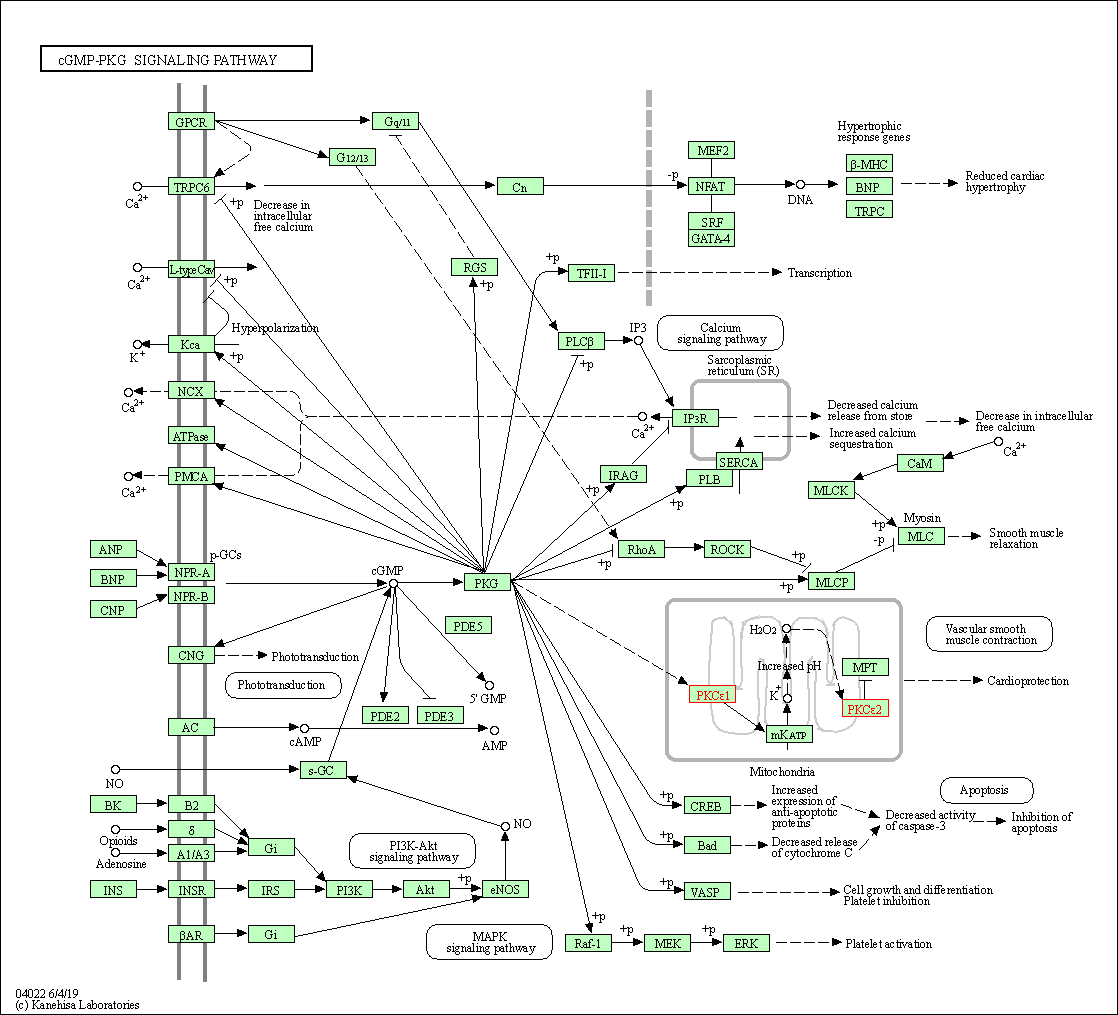
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |
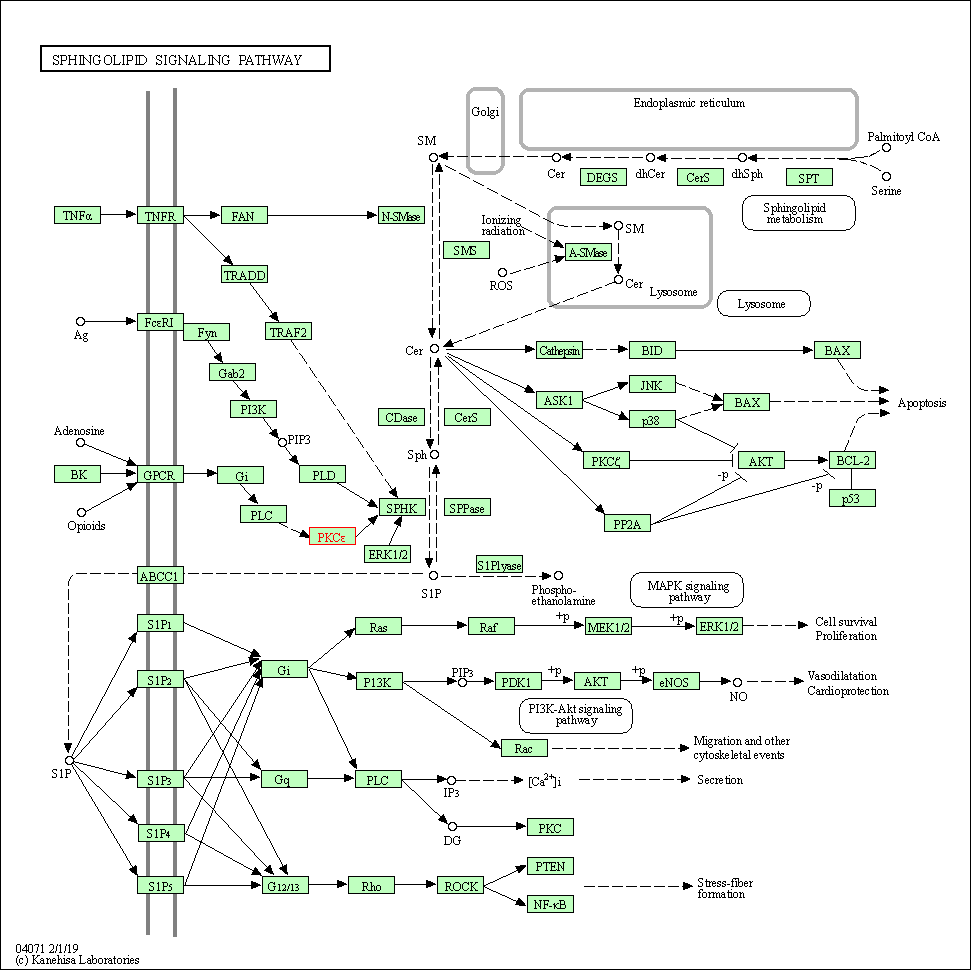
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |
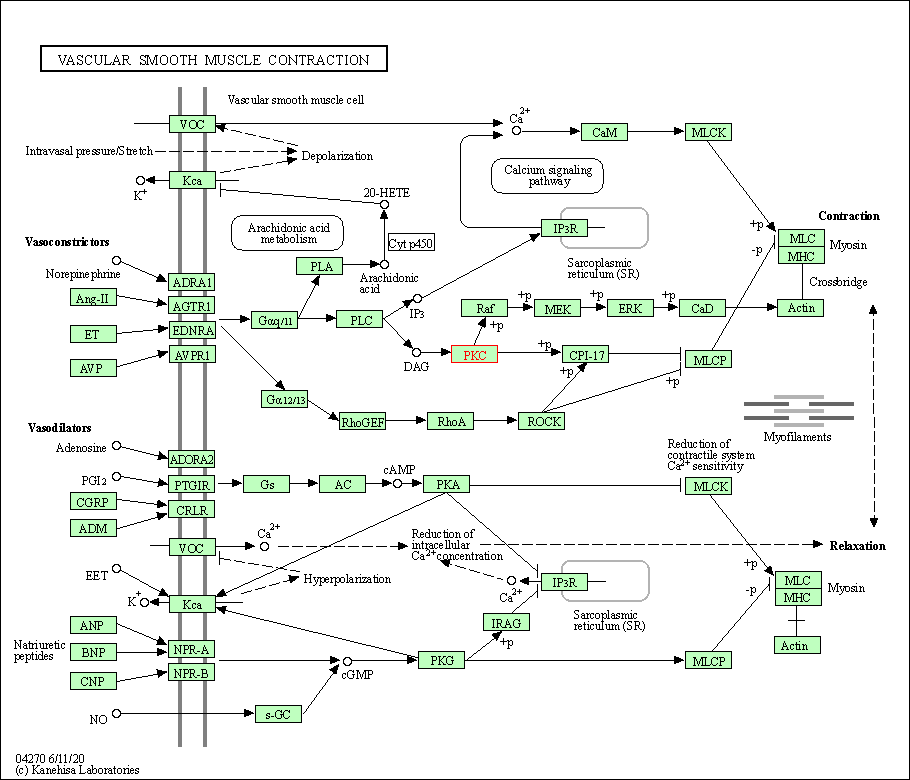
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Apelin signaling pathway | hsa04371 | Affiliated Target |
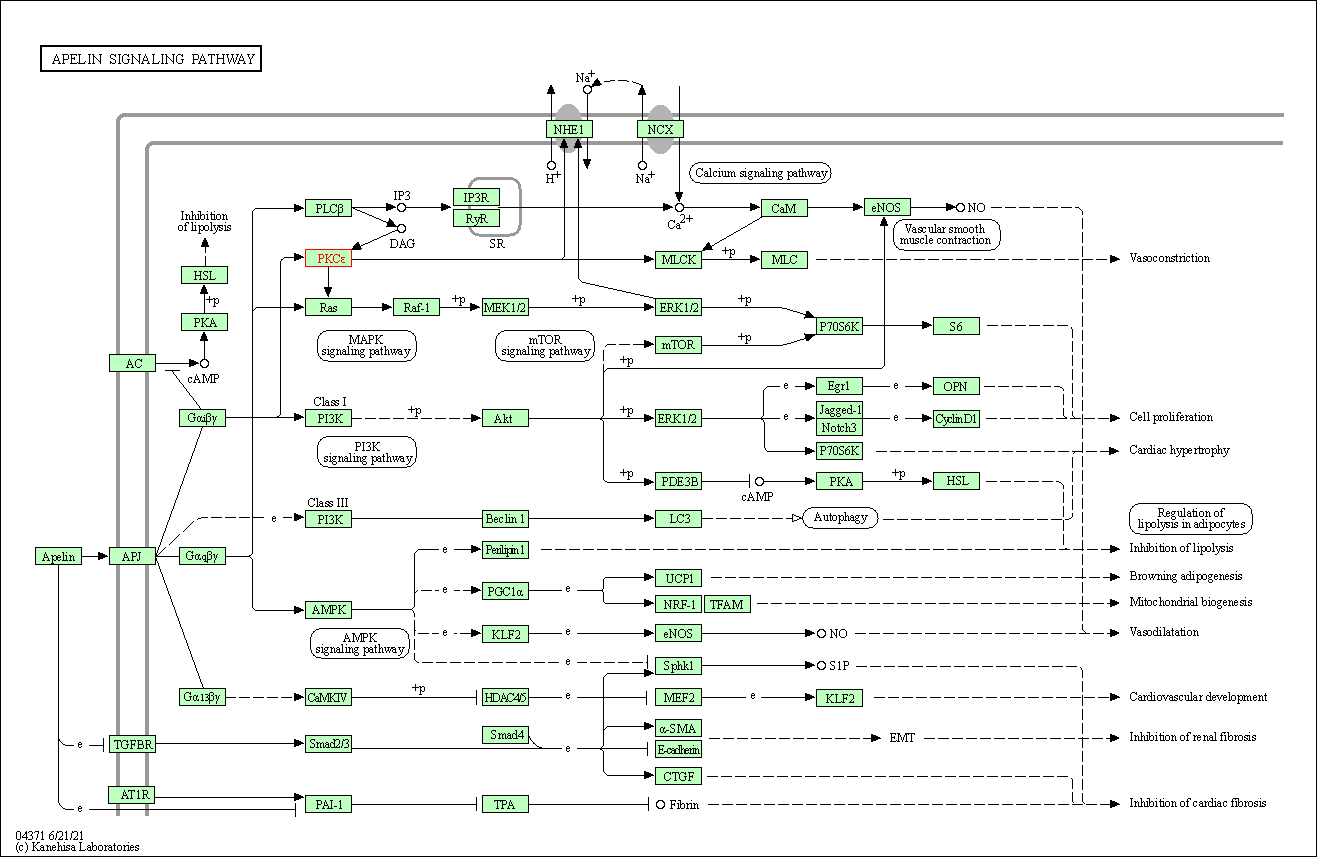
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Fc gamma R-mediated phagocytosis | hsa04666 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Aldosterone synthesis and secretion | hsa04925 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 9.03E-06 |
|---|---|---|---|---|---|
| Closeness centrality | 2.32E-01 | Radiality | 1.41E+01 | Clustering coefficient | 1.67E-01 |
| Neighborhood connectivity | 7.28E+01 | Topological coefficient | 2.73E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Protein kinase C, an elusive therapeutic target . Nat Rev Drug Discov. 2012 December; 11(12): 937-957. | |||||
| REF 2 | The fight against drug-resistant malaria: novel plasmodial targets and antimalarial drugs. Curr Med Chem. 2008;15(2):161-71. | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 5 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800025002) | |||||
| REF 7 | Protein kinase epsilon dampens the secretory response of model intestinal epithelia during ischemia. Surgery. 2001 Aug;130(2):310-8. | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800025002) | |||||
| REF 9 | Bisindolylmaleimide inhibitors of protein kinase C. Further conformational restriction of a tertiary amine side chain, Bioorg. Med. Chem. Lett. 4(11):1303-1308 (1994). | |||||
| REF 10 | Multivariate analysis by the minimum spanning tree method of the structural determinants of diphenylethylenes and triphenylacrylonitriles implicate... J Med Chem. 1992 Feb 7;35(3):573-83. | |||||
| REF 11 | Design, synthesis, and biological evaluation of 3,4-diarylmaleimides as angiogenesis inhibitors. J Med Chem. 2006 Feb 23;49(4):1271-81. | |||||
| REF 12 | (S)-13-[(dimethylamino)methyl]-10,11,14,15-tetrahydro-4,9:16, 21-dimetheno-1H, 13H-dibenzo[e,k]pyrrolo[3,4-h][1,4,13]oxadiazacyclohexadecene-1,3(2H... J Med Chem. 1996 Jul 5;39(14):2664-71. | |||||
| REF 13 | Inhibitors of protein kinase C. 1. 2,3-Bisarylmaleimides. J Med Chem. 1992 Jan;35(1):177-84. | |||||
| REF 14 | Novel protein kinase C inhibitors: synthesis and PKC inhibition of beta-substituted polythiophene derivatives. Bioorg Med Chem Lett. 1999 Aug 2;9(15):2279-82. | |||||
| REF 15 | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization. Dev Cell. 2016 Aug 22;38(4):384-98. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

