Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T02703
(Former ID: TTDS00337)
|
|||||
| Target Name |
Nitric-oxide synthase inducible (NOS2)
|
|||||
| Synonyms |
iNOS; Peptidyl-cysteine S-nitrosylase NOS2; Nitric oxide synthase, inducible; NOS2A; NOS type II; Inducible NOS; Inducible NO synthase; Hepatocyte NOS; HEP-NOS
Click to Show/Hide
|
|||||
| Gene Name |
NOS2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Osteoarthritis [ICD-11: FA00-FA05] | |||||
| 2 | Sepsis [ICD-11: 1G40-1G41] | |||||
| 3 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| 4 | Chronic kidney disease [ICD-11: GB61] | |||||
| Function |
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. In macrophages, NO mediates tumoricidal and bactericidal actions. Also has nitrosylase activity and mediates cysteine S-nitrosylation of cytoplasmic target proteins such PTGS2/COX2 (By similarity). As component of the iNOS-S100A8/9 transnitrosylase complex involved in the selective inflammatory stimulus-dependent S-nitrosylation of GAPDH on 'Cys-247' implicated in regulation of the GAIT complex activity and probably multiple targets including ANXA5, EZR, MSN and VIM. Involved in inflammation, enhances the synthesis of proinflammatory mediators such as IL6 and IL8.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.13.39
|
|||||
| Sequence |
MACPWKFLFKTKFHQYAMNGEKDINNNVEKAPCATSSPVTQDDLQYHNLSKQQNESPQPL
VETGKKSPESLVKLDATPLSSPRHVRIKNWGSGMTFQDTLHHKAKGILTCRSKSCLGSIM TPKSLTRGPRDKPTPPDELLPQAIEFVNQYYGSFKEAKIEEHLARVEAVTKEIETTGTYQ LTGDELIFATKQAWRNAPRCIGRIQWSNLQVFDARSCSTAREMFEHICRHVRYSTNNGNI RSAITVFPQRSDGKHDFRVWNAQLIRYAGYQMPDGSIRGDPANVEFTQLCIDLGWKPKYG RFDVVPLVLQANGRDPELFEIPPDLVLEVAMEHPKYEWFRELELKWYALPAVANMLLEVG GLEFPGCPFNGWYMGTEIGVRDFCDVQRYNILEEVGRRMGLETHKLASLWKDQAVVEINI AVLHSFQKQNVTIMDHHSAAESFMKYMQNEYRSRGGCPADWIWLVPPMSGSITPVFHQEM LNYVLSPFYYYQVEAWKTHVWQDEKRRPKRREIPLKVLVKAVLFACMLMRKTMASRVRVT ILFATETGKSEALAWDLGALFSCAFNPKVVCMDKYRLSCLEEERLLLVVTSTFGNGDCPG NGEKLKKSLFMLKELNNKFRYAVFGLGSSMYPRFCAFAHDIDQKLSHLGASQLTPMGEGD ELSGQEDAFRSWAVQTFKAACETFDVRGKQHIQIPKLYTSNVTWDPHHYRLVQDSQPLDL SKALSSMHAKNVFTMRLKSRQNLQSPTSSRATILVELSCEDGQGLNYLPGEHLGVCPGNQ PALVQGILERVVDGPTPHQTVRLEALDESGSYWVSDKRLPPCSLSQALTYFLDITTPPTQ LLLQKLAQVATEEPERQRLEALCQPSEYSKWKFTNSPTFLEVLEEFPSLRVSAGFLLSQL PILKPRFYSISSSRDHTPTEIHLTVAVVTYHTRDGQGPLHHGVCSTWLNSLKPQDPVPCF VRNASGFHLPEDPSHPCILIGPGTGIAPFRSFWQQRLHDSQHKGVRGGRMTLVFGCRRPD EDHIYQEEMLEMAQKGVLHAVHTAYSRLPGKPKVYVQDILRQQLASEVLRVLHKEPGHLY VCGDVRMARDVAHTLKQLVAAKLKLNEEQVEDYFFQLKSQKRYHEDIFGAVFPYEAKKDR VAVQPSSLEMSAL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A01796 ; BADD_A05438 | |||||
| HIT2.0 ID | T09H2Z | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 10 Clinical Trial Drugs | + | ||||
| 1 | Curcumin | Drug Info | Phase 3 | Solid tumour/cancer | [2], [3] | |
| 2 | MPL-S | Drug Info | Phase 3 | Endotoxic shock | [4] | |
| 3 | SD-6010 | Drug Info | Phase 3 | Osteoarthritis | [5] | |
| 4 | Pimagedine HCl | Drug Info | Phase 2/3 | Diabetic kidney disease | [6] | |
| 5 | BXT-51072 | Drug Info | Phase 2 | Cardiovascular disease | [7] | |
| 6 | CR-3294 | Drug Info | Phase 2 | Diarrhea | [8] | |
| 7 | GW274150 | Drug Info | Phase 2 | Asthma | [9] | |
| 8 | HP-228 | Drug Info | Phase 2 | Postoperative pain | [10] | |
| 9 | KD-7040 | Drug Info | Phase 2 | Pain | [11] | |
| 10 | LT-1951 | Drug Info | Phase 1/2 | Coronary artery disease | [12] | |
| Discontinued Drug(s) | [+] 2 Discontinued Drugs | + | ||||
| 1 | NOStentin | Drug Info | Discontinued in Phase 1 | Artery stenosis | [14] | |
| 2 | ONO-1714 | Drug Info | Discontinued in Phase 1 | Sepsis | [15] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 105 Inhibitor drugs | + | ||||
| 1 | Curcumin | Drug Info | [16] | |||
| 2 | SD-6010 | Drug Info | [17] | |||
| 3 | Pimagedine HCl | Drug Info | [18] | |||
| 4 | CR-3294 | Drug Info | [8], [20] | |||
| 5 | KD-7040 | Drug Info | [23] | |||
| 6 | ((E)-7-But-2-enyl)-azepan-(2Z)-ylideneamine | Drug Info | [28] | |||
| 7 | (4S,5R)-4,5-Dimethyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 8 | (4S,5R)-4,5-Dipropyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 9 | (4S,5S)-4,5-Diethyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 10 | (4S,5S)-4,5-Dipropyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 11 | (5-Imino-[1,4]thiazepan-3-yl)-methanol | Drug Info | [30] | |||
| 12 | (5S,6R)-[Octahydro-quinolin-(2E)-ylidene]amine | Drug Info | [31] | |||
| 13 | (5S,6S)-[Octahydro-quinolin-(2E)-ylidene]amine | Drug Info | [31] | |||
| 14 | (6r,1'r,2's)-5,6,7,8 Tetrahydrobiopterin | Drug Info | [32] | |||
| 15 | (R)-3-Propyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 16 | (S)-2-Amino-5-(N-methyl-guanidino)-pentanoic acid | Drug Info | [33] | |||
| 17 | (S)-3-Propyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 18 | (S)-6-Amino-2-(2-imino-ethylamino)-hexanoic acid | Drug Info | [30] | |||
| 19 | (S)-N-(1-phenylethyl)acetimidamide hydrobromide | Drug Info | [34] | |||
| 20 | 1-(6-Amino-4-methylpyridin-2-yl)propan-2-ol | Drug Info | [35] | |||
| 21 | 1400W | Drug Info | [34], [36], [37] | |||
| 22 | 2-(2-Amino-ethyl)-7-imino-azepane | Drug Info | [28] | |||
| 23 | 2-amino-4-methylpyridine | Drug Info | [38], [39] | |||
| 24 | 2-Amino-5-(N-nitro-guanidino)-pentanoic acid | Drug Info | [33] | |||
| 25 | 2-Aminothiazoline | Drug Info | [40] | |||
| 26 | 2-Methyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 27 | 3,4-Dihydro-1H-quinolin-(2E)-ylideneamine | Drug Info | [31] | |||
| 28 | 3,4-Dimethyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 29 | 3-(2-Amino-ethyl)-5-imino-[1,4]oxazepane | Drug Info | [28] | |||
| 30 | 3-(2-Nitro-ethyl)-[1,4]oxazepan-(5Z)-ylideneamine | Drug Info | [28] | |||
| 31 | 3-Bromo-1H-indazole-7-carbonitrile | Drug Info | [41] | |||
| 32 | 3-Butyl-[1,4]oxazepan-(5Z)-ylideneamine | Drug Info | [28] | |||
| 33 | 3-Butyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 34 | 3-Ethyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 35 | 3-Isobutyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 36 | 3-Methyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 37 | 3-Propyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 38 | 4,5,6,7-tetrafluoro-3-methyl-1H-indazole | Drug Info | [42] | |||
| 39 | 4,5-Dimethyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 40 | 4-Butyl-thiazolidin-(2E)-ylideneamine | Drug Info | [40] | |||
| 41 | 4-Ethyl-3-methyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 42 | 4-Ethyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 43 | 4-Ethyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 44 | 4-Isopropyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 45 | 4-Methyl-5-propyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 46 | 4-Methyl-6-(2-methylprop-1-enyl)pyridin-2-amine | Drug Info | [35] | |||
| 47 | 4-methyl-6-propylpyridin-2-amine | Drug Info | [39] | |||
| 48 | 4-Methyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 49 | 4-Methyl-piperidin-(2E)-ylideneamine | Drug Info | [31] | |||
| 50 | 4-Methyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 51 | 4-[(2-Methyl-1H-imidazol-1-yl)methyl]pyridine | Drug Info | [34] | |||
| 52 | 4r-Fluoro-N6-Ethanimidoyl-L-Lysine | Drug Info | [32] | |||
| 53 | 5-Bromomethyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 54 | 5-Ethyl-4-propyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 55 | 5-Ethyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 56 | 5-Methyl-4-propyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 57 | 5-Methyl-oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 58 | 5-Methyl-pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 59 | 6-(2-Fluoropropyl)-4-methylpyridin-2-amine | Drug Info | [35] | |||
| 60 | 6-(3-Fluoropropyl)-4-methylpyridin-2-amine | Drug Info | [35] | |||
| 61 | 6-(4-Fluorobutyl)-4-methylpyridin-2-amine | Drug Info | [35] | |||
| 62 | 6-isobutyl-4-methylpyridin-2-amine | Drug Info | [35] | |||
| 63 | 6-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine | Drug Info | [43] | |||
| 64 | 7,8-dihydrobiopterin | Drug Info | [44], [32] | |||
| 65 | 7-(2-Nitro-ethyl)-azepan-(2Z)-ylideneamine | Drug Info | [28] | |||
| 66 | 7-Butyl-azepan-(2Z)-ylideneamine | Drug Info | [28] | |||
| 67 | 7-Methyl-[1,4]thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 68 | 7-nitro-1H-indazole | Drug Info | [32], [42] | |||
| 69 | AR-C102222 | Drug Info | [39] | |||
| 70 | AR-C133057XX | Drug Info | [39] | |||
| 71 | Azepan-(2Z)-ylideneamine | Drug Info | [28] | |||
| 72 | Azocan-(2Z)-ylideneamine | Drug Info | [28] | |||
| 73 | Azonan-(2Z)-ylideneamine | Drug Info | [33] | |||
| 74 | B-Octylglucoside | Drug Info | [32] | |||
| 75 | BYK-191023 | Drug Info | [45] | |||
| 76 | CDDO | Drug Info | [46] | |||
| 77 | Ethylisothiourea | Drug Info | [32] | |||
| 78 | Hexahydro-cyclopenta[c]pyrrol-(1Z)-ylideneamine | Drug Info | [40] | |||
| 79 | KD-7332 | Drug Info | [45] | |||
| 80 | L-NIO | Drug Info | [47] | |||
| 81 | L-Thiocitrulline | Drug Info | [32] | |||
| 82 | N,N-dimethylarginine | Drug Info | [48] | |||
| 83 | N-benzylacetimidamide hydrobromide | Drug Info | [34] | |||
| 84 | N-omega-allyl-L-arginine | Drug Info | [47] | |||
| 85 | N-Omega-Hydroxy-L-Arginine | Drug Info | [32] | |||
| 86 | N-omega-propargyl-L-arginine | Drug Info | [47] | |||
| 87 | N-Omega-Propyl-L-Arginine | Drug Info | [32] | |||
| 88 | N5-(1-iminobut-3-enyl)-L-ornithine | Drug Info | [47] | |||
| 89 | N5-(1-iminobutyl)-L-ornithine | Drug Info | [47] | |||
| 90 | N5-(1-iminopropyl)-L-ornithine | Drug Info | [47] | |||
| 91 | Octahydro-isoindol-(1Z)-ylideneamine | Drug Info | [40] | |||
| 92 | Oxazolidin-(2Z)-ylideneamine | Drug Info | [29] | |||
| 93 | PIBTU | Drug Info | [49] | |||
| 94 | Piperidin-(2E)-ylideneamine | Drug Info | [31] | |||
| 95 | Pyrrolidin-(2Z)-ylideneamine | Drug Info | [40] | |||
| 96 | Resveratrol Potassium4,-Sulfate | Drug Info | [50] | |||
| 97 | THIOCITRULLINE | Drug Info | [51] | |||
| 98 | Thiocoumarin | Drug Info | [37] | |||
| 99 | [1,3]Oxazinan-(2E)-ylideneamine | Drug Info | [33] | |||
| 100 | [1,3]Thiazinan-(2E)-ylideneamine | Drug Info | [33] | |||
| 101 | [1,4]Oxazepan-(3E)-ylideneamine | Drug Info | [30] | |||
| 102 | [1,4]Oxazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 103 | [1,4]Thiazepan-(3E)-ylideneamine | Drug Info | [30] | |||
| 104 | [1,4]Thiazepan-(5E)-ylideneamine | Drug Info | [30] | |||
| 105 | [1,5]Thiazocan-(4E)-ylideneamine | Drug Info | [28] | |||
| Modulator | [+] 8 Modulator drugs | + | ||||
| 1 | MPL-S | Drug Info | [1] | |||
| 2 | BXT-51072 | Drug Info | [19] | |||
| 3 | GW274150 | Drug Info | [21], [22] | |||
| 4 | HP-228 | Drug Info | [10] | |||
| 5 | LT-1951 | Drug Info | [24] | |||
| 6 | NOStentin | Drug Info | [25] | |||
| 7 | ONO-1714 | Drug Info | [26], [27] | |||
| 8 | SKLB-010 | Drug Info | [45] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Arginine | Ligand Info | |||||
| Structure Description | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | PDB:1NSI | ||||
| Method | X-ray diffraction | Resolution | 2.55 Å | Mutation | No | [52] |
| PDB Sequence |
RHVRIKNWGS
92 GMTFQDTLHH102 KAKGILTCRS112 KSCLGSIMTP122 KSLTRGPRDK132 PTPPDELLPQ 142 AIEFVNQYYG152 SFKEAKIEEH162 LARVEAVTKE172 IETTGTYQLT182 GDELIFATKQ 192 AWRNAPRCIG202 RIQWSNLQVF212 DARSCSTARE222 MFEHICRHVR232 YSTNNGNIRS 242 AITVFPQRSD252 GKHDFRVWNA262 QLIRYAGYQM272 PDGSIRGDPA282 NVEFTQLCID 292 LGWKPKYGRF302 DVVPLVLQAN312 GRDPELFEIP322 PDLVLEVAME332 HPKYEWFREL 342 ELKWYALPAV352 ANMLLEVGGL362 EFPGCPFNGW372 YMGTEIGVRD382 FCDVQRYNIL 392 EEVGRRMGLE402 THKLASLWKD412 QAVVEINIAV422 LHSFQKQNVT432 IMDHHSAAES 442 FMKYMQNEYR452 SRGGCPADWI462 WLVPPMSGSI472 TPVFHQEMLN482 YVLSPFYYYQ 492 VEAWKTHVWQ502
|
|||||
|
|
||||||
| Ligand Name: Ethylisothiourea | Ligand Info | |||||
| Structure Description | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE WITH INHIBITOR | PDB:4NOS | ||||
| Method | X-ray diffraction | Resolution | 2.25 Å | Mutation | No | [53] |
| PDB Sequence |
RHVRIKNWGS
92 GMTFQDTLHH102 KAKGILTCRS112 KSCLGSIMTP122 KSLTRGPRDK132 PTPPDELLPQ 142 AIEFVNQYYG152 SFKEAKIEEH162 LARVEAVTKE172 IETTGTYQLT182 GDELIFATKQ 192 AWRNAPRCIG202 RIQWSNLQVF212 DARSCSTARE222 MFEHICRHVR232 YSTNNGNIRS 242 AITVFPQRSD252 GKHDFRVWNA262 QLIRYAGYQM272 PDGSIRGDPA282 NVEFTQLCID 292 LGWKPKYGRF302 DVVPLVLQAN312 GRDPELFEIP322 PDLVLEVAME332 HPKYEWFREL 342 ELKWYALPAV352 ANMLLEVGGL362 EFPGCPFNGW372 YMGTEIGVRD382 FCDVQRYNIL 392 EEVGRRMGLE402 THKLASLWKD412 QAVVEINIAV422 IHSFQKQNVT432 IMDHHSAAES 442 FMKYMQNEYR452 SRGGCPADWI462 WLVPPMSGSI472 TPVFHQEMLN482 YVLSPFYYYQ 492 VEAWKTHVWQ502 D
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Arginine biosynthesis | hsa00220 | Affiliated Target |
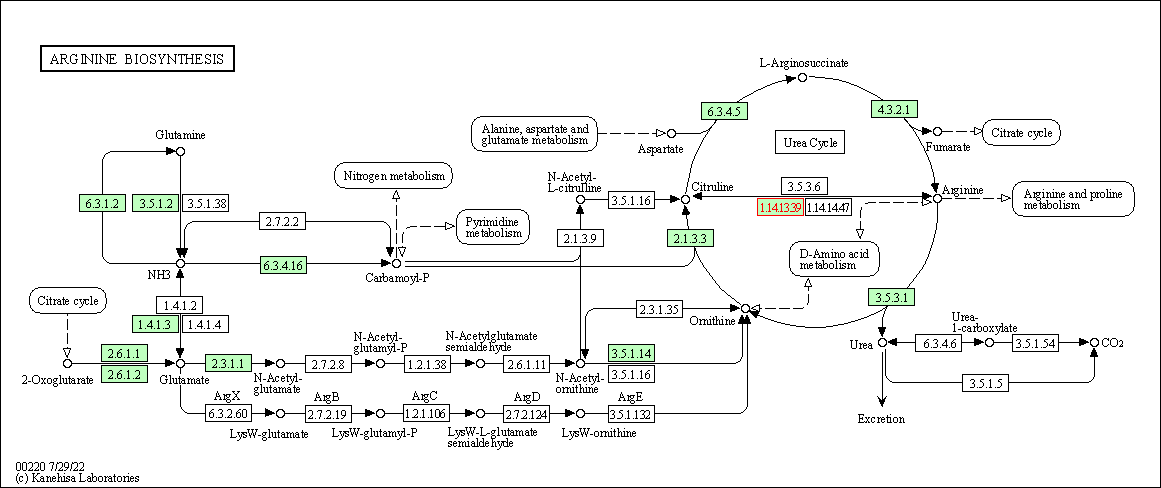
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Arginine and proline metabolism | hsa00330 | Affiliated Target |
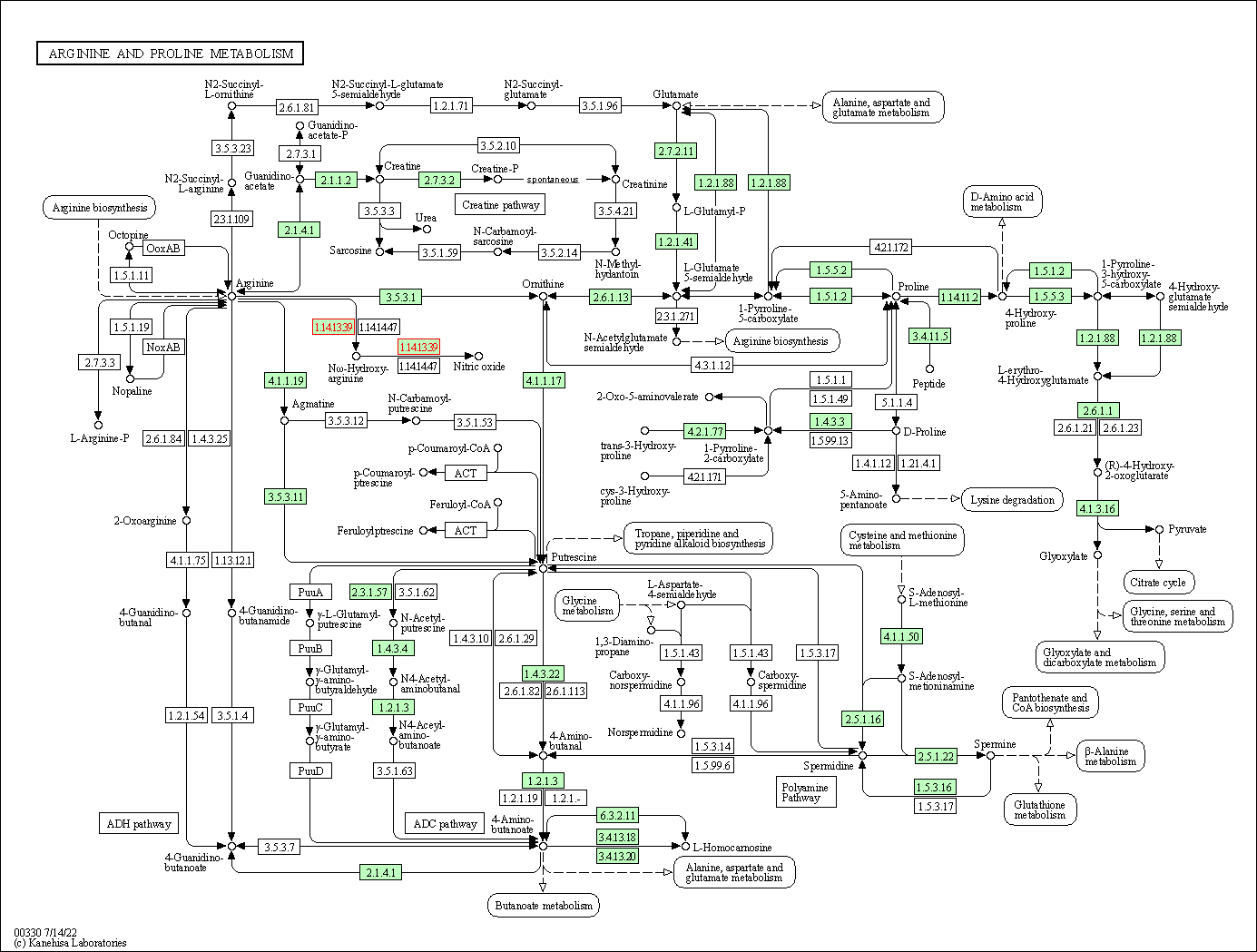
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Calcium signaling pathway | hsa04020 | Affiliated Target |
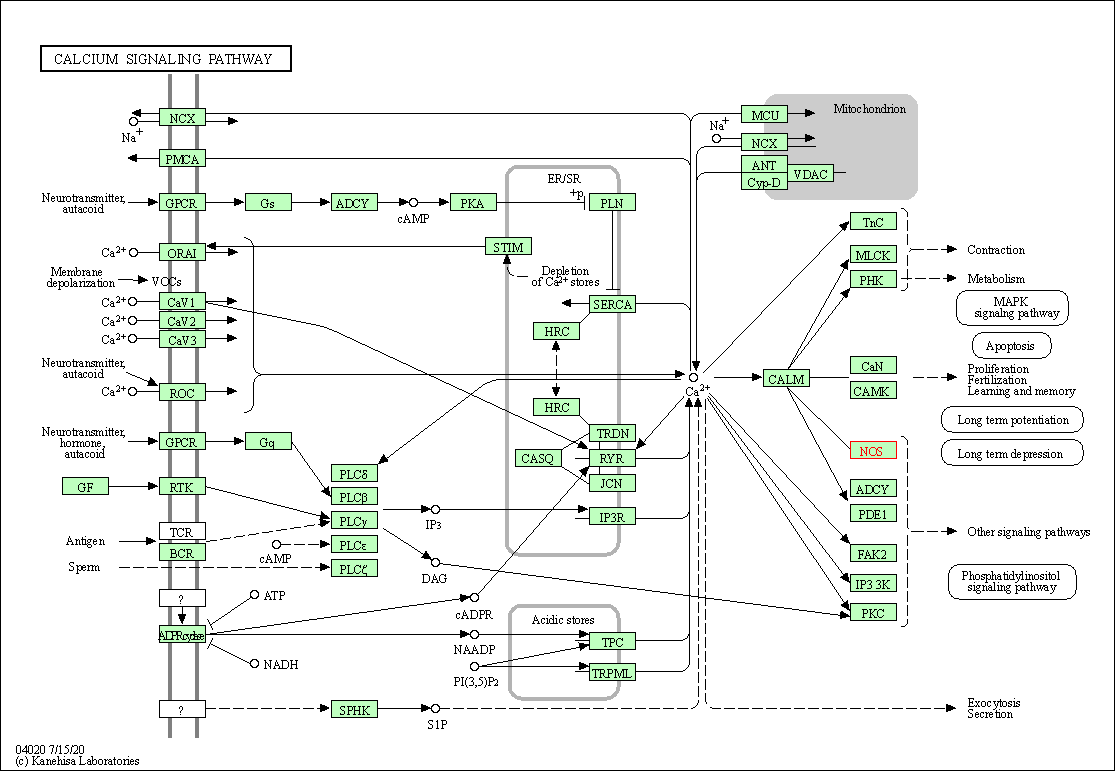
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |
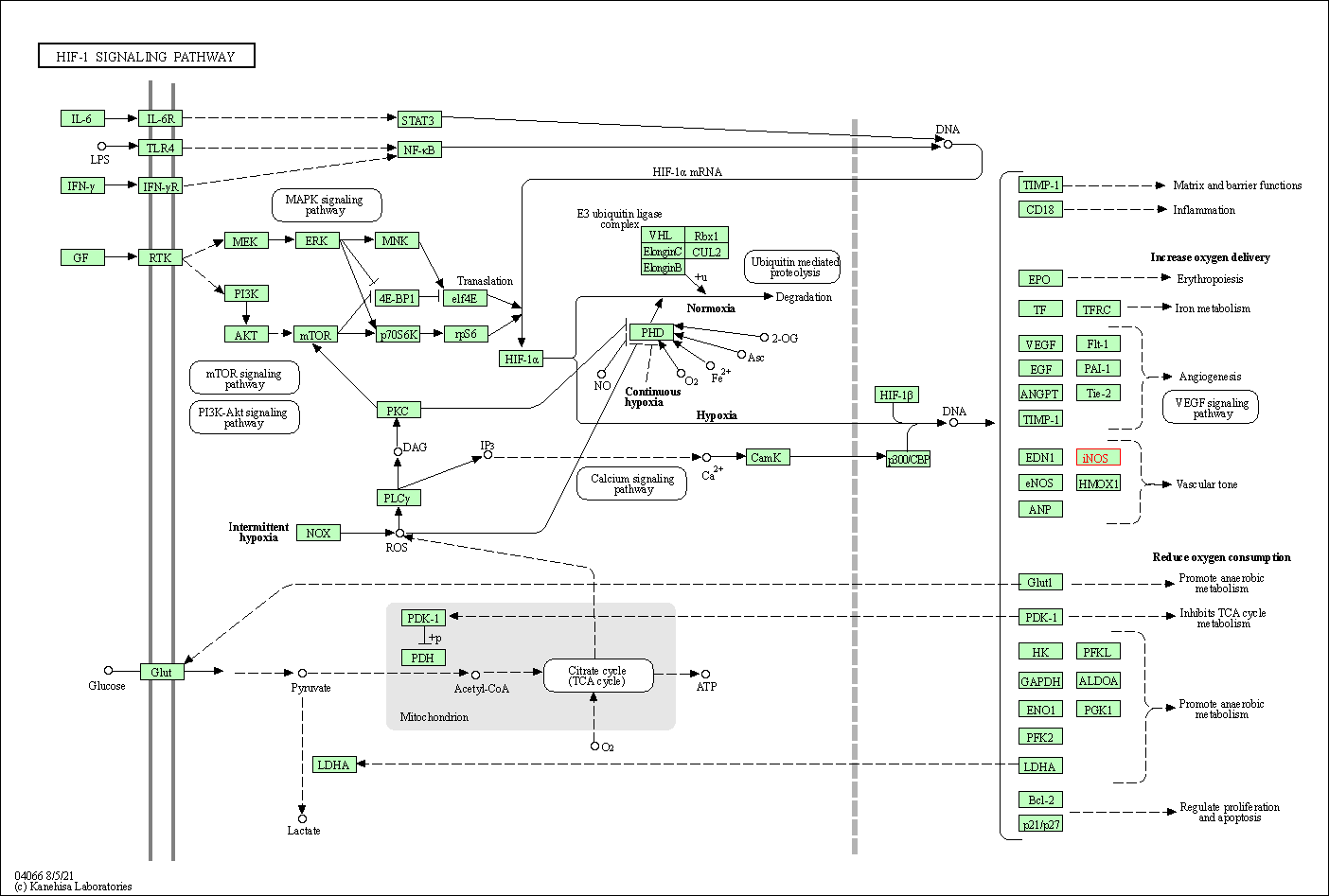
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Peroxisome | hsa04146 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Apelin signaling pathway | hsa04371 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 12 | Degree centrality | 1.29E-03 | Betweenness centrality | 2.14E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.48E-01 | Radiality | 1.43E+01 | Clustering coefficient | 1.67E-01 |
| Neighborhood connectivity | 6.26E+01 | Topological coefficient | 1.18E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Lipopolysaccharide and monophosphoryl lipid A differentially regulate interleukin-12, gamma interferon, and interleukin-10 mRNA production in murine macrophages. Infect Immun. 1997 Aug;65(8):3239-47. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7000). | |||||
| REF 3 | Nanocurcumin: a promising therapeutic advancement over native curcumin. Crit Rev Ther Drug Carrier Syst. 2013;30(4):331-68. | |||||
| REF 4 | ClinicalTrials.gov (NCT00699764) Safety of a Herpes Simplex Candidate Vaccine (gD2t) With MPL and Its Efficacy to Prevent Genital Herpes Disease. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT01438918) X-Ray Study Investigating The Safety And Efficacy Of SD-6010 In Subjects With Osteoarthritis Of The Knee. U.S. National Institutes of Health. | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800006816) | |||||
| REF 7 | ClinicalTrials.gov (NCT00782613) Efficacy and Safety of Topical ALT-2074 (SYI-2074) for Treatment of Chronic Plaque Psoriasis. U.S. National Institutes of Health. | |||||
| REF 8 | Efficacy of CR3294, a new benzamidine derivative, in the prevention of 5-fluorouracil-induced gastrointestinal mucositis and diarrhea in mice. Cancer Chemother Pharmacol. 2010 Oct;66(5):819-27. | |||||
| REF 9 | ClinicalTrials.gov (NCT00370435) Study Of 90mg Of GW274150 In Subjects Over 50 Years, Who Have Rheumatoid Arthritis (RA). U.S. National Institutes of Health. | |||||
| REF 10 | HP-228, a novel synthetic peptide, inhibits the induction of nitric oxide synthase in vivo but not in vitro. J Pharmacol Exp Ther. 1995 Nov;275(2):584-91. | |||||
| REF 11 | ClinicalTrials.gov (NCT00576108) A 2 Week Study of Topical KD7040 in the Treatment of Postherpetic Neuralgia (PHN). U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT00264706) PolyArginine Treated vEiN grafTs (PATENT). U.S. National Institutes of Health. | |||||
| REF 13 | ClinicalTrials.gov (NCT02099981) Restoration of Retinal Vascular Responses in Type 1 Diabetic Patients in University of Minnesota - Clinical and Translational Science Institute. | |||||
| REF 14 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800013241) | |||||
| REF 15 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800013758) | |||||
| REF 16 | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20523-8. | |||||
| REF 17 | A 2-year randomised, double-blind, placebo-controlled, multicentre study of oral selective iNOS inhibitor, cindunistat (SD-6010), in patients with symptomatic osteoarthritis of the knee. Ann Rheum Dis. 2013 Feb;72(2):187-95. | |||||
| REF 18 | Nitric oxide associated with iNOS expression inhibits acetylcholinesterase activity and induces memory impairment during acute hypobaric hypoxia. Brain Res. 2008 Sep 16;1230:138-49. | |||||
| REF 19 | Shear stress induces iNOS expression in cultured smooth muscle cells: role of oxidative stress. Am J Physiol Cell Physiol. 2000 Dec;279(6):C1880-8. | |||||
| REF 20 | Efficacy of CR3294, a new benzamidine derivative, in the prevention of 5-fluorouracil-induced gastrointestinal mucositis and diarrhea in mice. Cancer Chemother Pharmacol. 2010 October; 66(5): 819-827. | |||||
| REF 21 | GW274150, a potent and highly selective inhibitor of iNOS, reduces experimental renal ischemia/reperfusion injury. Kidney Int. 2003 Mar;63(3):853-65. | |||||
| REF 22 | GW274150 and GW273629 are potent and highly selective inhibitors of inducible nitric oxide synthase in vitro and in vivo. Br J Pharmacol. 2005 Jun;145(3):301-12. | |||||
| REF 23 | WO patent application no. 2014,0214,08, Method for treating cancer by anticancer agent co-administration. | |||||
| REF 24 | Critical role of L-arginine in endothelial cell survival during oxidative stress. Circulation. 2003 May 27;107(20):2607-14. | |||||
| REF 25 | Gene therapy with iNOS provides long-term protection against myocardial infarction without adverse functional consequences. Am J Physiol Heart Circ Physiol. 2006 Feb;290(2):H584-9. | |||||
| REF 26 | ONO-1714, a new inducible nitric oxide synthase inhibitor, attenuates diaphragmatic dysfunction associated with cerulein-induced pancreatitis in rats. Crit Care Med. 2001 Jun;29(6):1215-21. | |||||
| REF 27 | The potent inducible nitric oxide synthase inhibitor ONO-1714 inhibits neuronal NOS and exerts antinociception in rats. Neurosci Lett. 2004 Jul 22;365(2):111-5. | |||||
| REF 28 | Selective heterocyclic amidine inhibitors of human inducible nitric oxide synthase. Bioorg Med Chem Lett. 2001 Oct 8;11(19):2651-3. | |||||
| REF 29 | 4,5-Disubstituted-1,3-oxazolidin-2-imine derivatives: a new class of orally bioavailable nitric oxide synthase inhibitor. Bioorg Med Chem Lett. 2004 Jan 19;14(2):313-6. | |||||
| REF 30 | Synthesis of analogs of (1,4)-3- and 5-imino oxazepane, thiazepane, and diazepane as inhibitors of nitric oxide synthases. Bioorg Med Chem Lett. 2004 Dec 6;14(23):5907-11. | |||||
| REF 31 | Bicyclic amidine inhibitors of nitric oxide synthase: discovery of perhydro-iminopyrindine and perhydro-iminoquinoline as potent, orally active inh... Bioorg Med Chem Lett. 2005 Apr 15;15(8):1997-2001. | |||||
| REF 32 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 33 | 2-Iminopiperidine and other 2-iminoazaheterocycles as potent inhibitors of human nitric oxide synthase isoforms. J Med Chem. 1996 Feb 2;39(3):669-72. | |||||
| REF 34 | N-Substituted acetamidines and 2-methylimidazole derivatives as selective inhibitors of neuronal nitric oxide synthase. Bioorg Med Chem Lett. 2010 Nov 15;20(22):6495-9. | |||||
| REF 35 | Design and synthesis of 2-amino-4-methylpyridine analogues as inhibitors for inducible nitric oxide synthase and in vivo evaluation of [18F]6-(2-fl... J Med Chem. 2009 Apr 23;52(8):2443-53. | |||||
| REF 36 | 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J Biol Chem. 1997 Feb 21;272(8):4959-63. | |||||
| REF 37 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 38 | 2-Amino-4-methylpyridine as a potent inhibitor of inducible NO synthase activity in vitro and in vivo. Br J Pharmacol. 1996 Nov;119(6):1101-8. | |||||
| REF 39 | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat Chem Biol. 2008 Nov;4(11):700-7. | |||||
| REF 40 | Evaluation of pyrrolidin-2-imines and 1,3-thiazolidin-2-imines as inhibitors of nitric oxide synthase. Bioorg Med Chem Lett. 2004 Sep 6;14(17):4539-44. | |||||
| REF 41 | Inhibitory effects of a series of 7-substituted-indazoles toward nitric oxide synthases: particular potency of 1H-indazole-7-carbonitrile. Bioorg Med Chem. 2008 Jun 1;16(11):5962-73. | |||||
| REF 42 | Fluorinated indazoles as novel selective inhibitors of nitric oxide synthase (NOS): synthesis and biological evaluation. Bioorg Med Chem. 2009 Sep 1;17(17):6180-7. | |||||
| REF 43 | Synthesis of biflavones having a 6-O-7'' linkage and effects on cyclooxygenase-2 and inducible nitric oxide synthase. Bioorg Med Chem Lett. 2009 Jan 1;19(1):74-6. | |||||
| REF 44 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 45 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1250). | |||||
| REF 46 | Studies on the reactivity of CDDO, a promising new chemopreventive and chemotherapeutic agent: implications for a molecular mechanism of action. Bioorg Med Chem Lett. 2005 May 2;15(9):2215-9. | |||||
| REF 47 | Structure-activity relationship of novel and known inhibitors of human dimethylarginine dimethylaminohydrolase-1: alkenyl-amidines as new leads. Bioorg Med Chem. 2008 Dec 15;16(24):10205-9. | |||||
| REF 48 | Homocysteine and asymmetric dimethylarginine (ADMA): biochemically linked but differently related to vascular disease in chronic kidney disease. Clin Chem Lab Med. 2007;45(12):1683-7. | |||||
| REF 49 | Potent and selective inhibition of human nitric oxide synthases. Inhibition by non-amino acid isothioureas. J Biol Chem. 1994 Oct 28;269(43):26669-76. | |||||
| REF 50 | Selective synthesis and biological evaluation of sulfate-conjugated resveratrol metabolites. J Med Chem. 2010 Jul 8;53(13):5033-43. | |||||
| REF 51 | Evaluation of 3-substituted arginine analogs as selective inhibitors of human nitric oxide synthase isozymes. Bioorg Med Chem Lett. 2005 Jun 2;15(11):2881-5. | |||||
| REF 52 | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase. J Biol Chem. 1999 Jul 23;274(30):21276-84. | |||||
| REF 53 | Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Biol. 1999 Mar;6(3):233-42. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

