Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T07533
(Former ID: TTDC00331)
|
|||||
| Target Name |
Apolipoprotein B-100 (APOB)
|
|||||
| Synonyms |
Apolipoprotein B48; Apo B48; Apo B100; Apo B-100
Click to Show/Hide
|
|||||
| Gene Name |
APOB
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Hyper-lipoproteinaemia [ICD-11: 5C80] | |||||
| Function |
Apo B-100 functions as a recognition signal for the cellular binding and internalization of LDL particles by the apoB/E receptor. Apolipoprotein B is a major protein constituent of chylomicrons (apo B-48), LDL (apo B-100) and VLDL (apo B-100).
Click to Show/Hide
|
|||||
| BioChemical Class |
Apolipoprotein
|
|||||
| UniProt ID | ||||||
| Sequence |
MDPPRPALLALLALPALLLLLLAGARAEEEMLENVSLVCPKDATRFKHLRKYTYNYEAES
SSGVPGTADSRSATRINCKVELEVPQLCSFILKTSQCTLKEVYGFNPEGKALLKKTKNSE EFAAAMSRYELKLAIPEGKQVFLYPEKDEPTYILNIKRGIISALLVPPETEEAKQVLFLD TVYGNCSTHFTVKTRKGNVATEISTERDLGQCDRFKPIRTGISPLALIKGMTRPLSTLIS SSQSCQYTLDAKRKHVAEAICKEQHLFLPFSYKNKYGMVAQVTQTLKLEDTPKINSRFFG EGTKKMGLAFESTKSTSPPKQAEAVLKTLQELKKLTISEQNIQRANLFNKLVTELRGLSD EAVTSLLPQLIEVSSPITLQALVQCGQPQCSTHILQWLKRVHANPLLIDVVTYLVALIPE PSAQQLREIFNMARDQRSRATLYALSHAVNNYHKTNPTGTQELLDIANYLMEQIQDDCTG DEDYTYLILRVIGNMGQTMEQLTPELKSSILKCVQSTKPSLMIQKAAIQALRKMEPKDKD QEVLLQTFLDDASPGDKRLAAYLMLMRSPSQADINKIVQILPWEQNEQVKNFVASHIANI LNSEELDIQDLKKLVKEALKESQLPTVMDFRKFSRNYQLYKSVSLPSLDPASAKIEGNLI FDPNNYLPKESMLKTTLTAFGFASADLIEIGLEGKGFEPTLEALFGKQGFFPDSVNKALY WVNGQVPDGVSKVLVDHFGYTKDDKHEQDMVNGIMLSVEKLIKDLKSKEVPEARAYLRIL GEELGFASLHDLQLLGKLLLMGARTLQGIPQMIGEVIRKGSKNDFFLHYIFMENAFELPT GAGLQLQISSSGVIAPGAKAGVKLEVANMQAELVAKPSVSVEFVTNMGIIIPDFARSGVQ MNTNFFHESGLEAHVALKAGKLKFIIPSPKRPVKLLSGGNTLHLVSTTKTEVIPPLIENR QSWSVCKQVFPGLNYCTSGAYSNASSTDSASYYPLTGDTRLELELRPTGEIEQYSVSATY ELQREDRALVDTLKFVTQAEGAKQTEATMTFKYNRQSMTLSSEVQIPDFDVDLGTILRVN DESTEGKTSYRLTLDIQNKKITEVALMGHLSCDTKEERKIKGVISIPRLQAEARSEILAH WSPAKLLLQMDSSATAYGSTVSKRVAWHYDEEKIEFEWNTGTNVDTKKMTSNFPVDLSDY PKSLHMYANRLLDHRVPQTDMTFRHVGSKLIVAMSSWLQKASGSLPYTQTLQDHLNSLKE FNLQNMGLPDFHIPENLFLKSDGRVKYTLNKNSLKIEIPLPFGGKSSRDLKMLETVRTPA LHFKSVGFHLPSREFQVPTFTIPKLYQLQVPLLGVLDLSTNVYSNLYNWSASYSGGNTST DHFSLRARYHMKADSVVDLLSYNVQGSGETTYDHKNTFTLSYDGSLRHKFLDSNIKFSHV EKLGNNPVSKGLLIFDASSSWGPQMSASVHLDSKKKQHLFVKEVKIDGQFRVSSFYAKGT YGLSCQRDPNTGRLNGESNLRFNSSYLQGTNQITGRYEDGTLSLTSTSDLQSGIIKNTAS LKYENYELTLKSDTNGKYKNFATSNKMDMTFSKQNALLRSEYQADYESLRFFSLLSGSLN SHGLELNADILGTDKINSGAHKATLRIGQDGISTSATTNLKCSLLVLENELNAELGLSGA SMKLTTNGRFREHNAKFSLDGKAALTELSLGSAYQAMILGVDSKNIFNFKVSQEGLKLSN DMMGSYAEMKFDHTNSLNIAGLSLDFSSKLDNIYSSDKFYKQTVNLQLQPYSLVTTLNSD LKYNALDLTNNGKLRLEPLKLHVAGNLKGAYQNNEIKHIYAISSAALSASYKADTVAKVQ GVEFSHRLNTDIAGLASAIDMSTNYNSDSLHFSNVFRSVMAPFTMTIDAHTNGNGKLALW GEHTGQLYSKFLLKAEPLAFTFSHDYKGSTSHHLVSRKSISAALEHKVSALLTPAEQTGT WKLKTQFNNNEYSQDLDAYNTKDKIGVELTGRTLADLTLLDSPIKVPLLLSEPINIIDAL EMRDAVEKPQEFTIVAFVKYDKNQDVHSINLPFFETLQEYFERNRQTIIVVLENVQRNLK HINIDQFVRKYRAALGKLPQQANDYLNSFNWERQVSHAKEKLTALTKKYRITENDIQIAL DDAKINFNEKLSQLQTYMIQFDQYIKDSYDLHDLKIAIANIIDEIIEKLKSLDEHYHIRV NLVKTIHDLHLFIENIDFNKSGSSTASWIQNVDTKYQIRIQIQEKLQQLKRHIQNIDIQH LAGKLKQHIEAIDVRVLLDQLGTTISFERINDILEHVKHFVINLIGDFEVAEKINAFRAK VHELIERYEVDQQIQVLMDKLVELAHQYKLKETIQKLSNVLQQVKIKDYFEKLVGFIDDA VKKLNELSFKTFIEDVNKFLDMLIKKLKSFDYHQFVDETNDKIREVTQRLNGEIQALELP QKAEALKLFLEETKATVAVYLESLQDTKITLIINWLQEALSSASLAHMKAKFRETLEDTR DRMYQMDIQQELQRYLSLVGQVYSTLVTYISDWWTLAAKNLTDFAEQYSIQDWAKRMKAL VEQGFTVPEIKTILGTMPAFEVSLQALQKATFQTPDFIVPLTDLRIPSVQINFKDLKNIK IPSRFSTPEFTILNTFHIPSFTIDFVEMKVKIIRTIDQMLNSELQWPVPDIYLRDLKVED IPLARITLPDFRLPEIAIPEFIIPTLNLNDFQVPDLHIPEFQLPHISHTIEVPTFGKLYS ILKIQSPLFTLDANADIGNGTTSANEAGIAASITAKGESKLEVLNFDFQANAQLSNPKIN PLALKESVKFSSKYLRTEHGSEMLFFGNAIEGKSNTVASLHTEKNTLELSNGVIVKINNQ LTLDSNTKYFHKLNIPKLDFSSQADLRNEIKTLLKAGHIAWTSSGKGSWKWACPRFSDEG THESQISFTIEGPLTSFGLSNKINSKHLRVNQNLVYESGSLNFSKLEIQSQVDSQHVGHS VLTAKGMALFGEGKAEFTGRHDAHLNGKVIGTLKNSLFFSAQPFEITASTNNEGNLKVRF PLRLTGKIDFLNNYALFLSPSAQQASWQVSARFNQYKYNQNFSAGNNENIMEAHVGINGE ANLDFLNIPLTIPEMRLPYTIITTPPLKDFSLWEKTGLKEFLKTTKQSFDLSVKAQYKKN KHRHSITNPLAVLCEFISQSIKSFDRHFEKNRNNALDFVTKSYNETKIKFDKYKAEKSHD ELPRTFQIPGYTVPVVNVEVSPFTIEMSAFGYVFPKAVSMPSFSILGSDVRVPSYTLILP SLELPVLHVPRNLKLSLPDFKELCTISHIFIPAMGNITYDFSFKSSVITLNTNAELFNQS DIVAHLLSSSSSVIDALQYKLEGTTRLTRKRGLKLATALSLSNKFVEGSHNSTVSLTTKN MEVSVATTTKAQIPILRMNFKQELNGNTKSKPTVSSSMEFKYDFNSSMLYSTAKGAVDHK LSLESLTSYFSIESSTKGDVKGSVLSREYSGTIASEANTYLNSKSTRSSVKLQGTSKIDD IWNLEVKENFAGEATLQRIYSLWEHSTKNHLQLEGLFFTNGEHTSKATLELSPWQMSALV QVHASQPSSFHDFPDLGQEVALNANTKNQKIRWKNEVRIHSGSFQSQVELSNDQEKAHLD IAGSLEGHLRFLKNIILPVYDKSLWDFLKLDVTTSIGRRQHLRVSTAFVYTKNPNGYSFS IPVKVLADKFIIPGLKLNDLNSVLVMPTFHVPFTDLQVPSCKLDFREIQIYKKLRTSSFA LNLPTLPEVKFPEVDVLTKYSQPEDSLIPFFEITVPESQLTVSQFTLPKSVSDGIAALDL NAVANKIADFELPTIIVPEQTIEIPSIKFSVPAGIVIPSFQALTARFEVDSPVYNATWSA SLKNKADYVETVLDSTCSSTVQFLEYELNVLGTHKIEDGTLASKTKGTFAHRDFSAEYEE DGKYEGLQEWEGKAHLNIKSPAFTDLHLRYQKDKKGISTSAASPAVGTVGMDMDEDDDFS KWNFYYSPQSSPDKKLTIFKTELRVRESDEETQIKVNWEEEAASGLLTSLKDNVPKATGV LYDYVNKYHWEHTGLTLREVSSKLRRNLQNNAEWVYQGAIRQIDDIDVRFQKAASGTTGT YQEWKDKAQNLYQELLTQEGQASFQGLKDNVFDGLVRVTQEFHMKVKHLIDSLIDFLNFP RFQFPGKPGIYTREELCTMFIREVGTVLSQVYSKVHNGSEILFSYFQDLVITLPFELRKH KLIDVISMYRELLKDLSKEAQEVFKAIQSLKTTEVLRNLQDLLQFIFQLIEDNIKQLKEM KFTYLINYIQDEINTIFSDYIPYVFKLLKENLCLNLHKFNEFIQNELQEASQELQQIHQY IMALREEYFDPSIVGWTVKYYELEEKIVSLIKNLLVALKDFHSEYIVSASNFTSQLSSQV EQFLHRNIQEYLSILTDPDGKGKEKIAELSATAQEIIKSQAIATKKIISDYHQQFRYKLQ DFSDQLSDYYEKFIAESKRLIDLSIQNYHTFLIYITELLKKLQSTTVMNPYMKLAPGELT IIL Click to Show/Hide
|
|||||
| ADReCS ID | BADD_A02584 | |||||
| HIT2.0 ID | T73S71 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Periaxin (PRX) | 29.508 (54/183) | 9.37E-09 | |
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Fat digestion and absorption | hsa04975 | Affiliated Target |
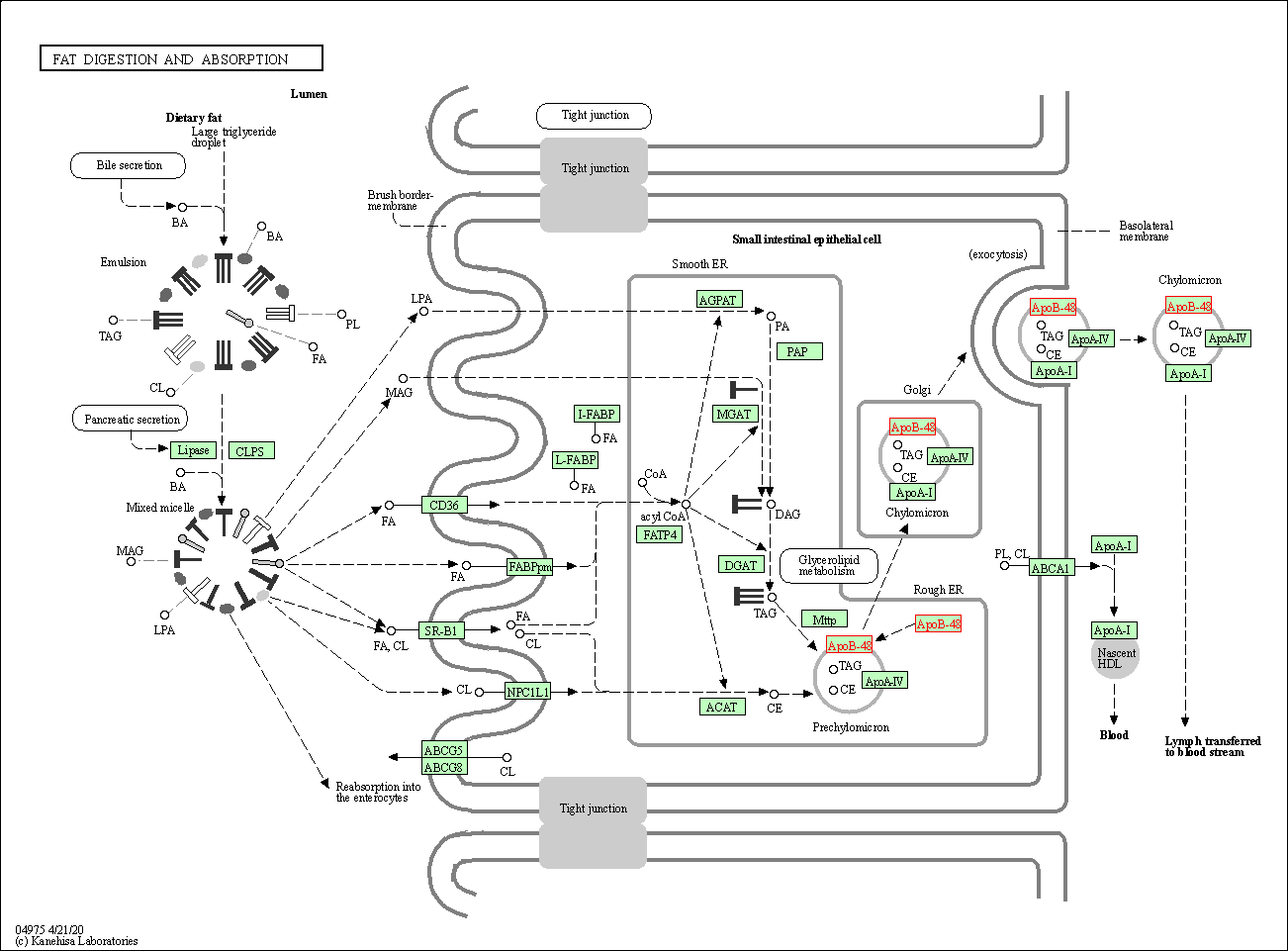
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Vitamin digestion and absorption | hsa04977 | Affiliated Target |
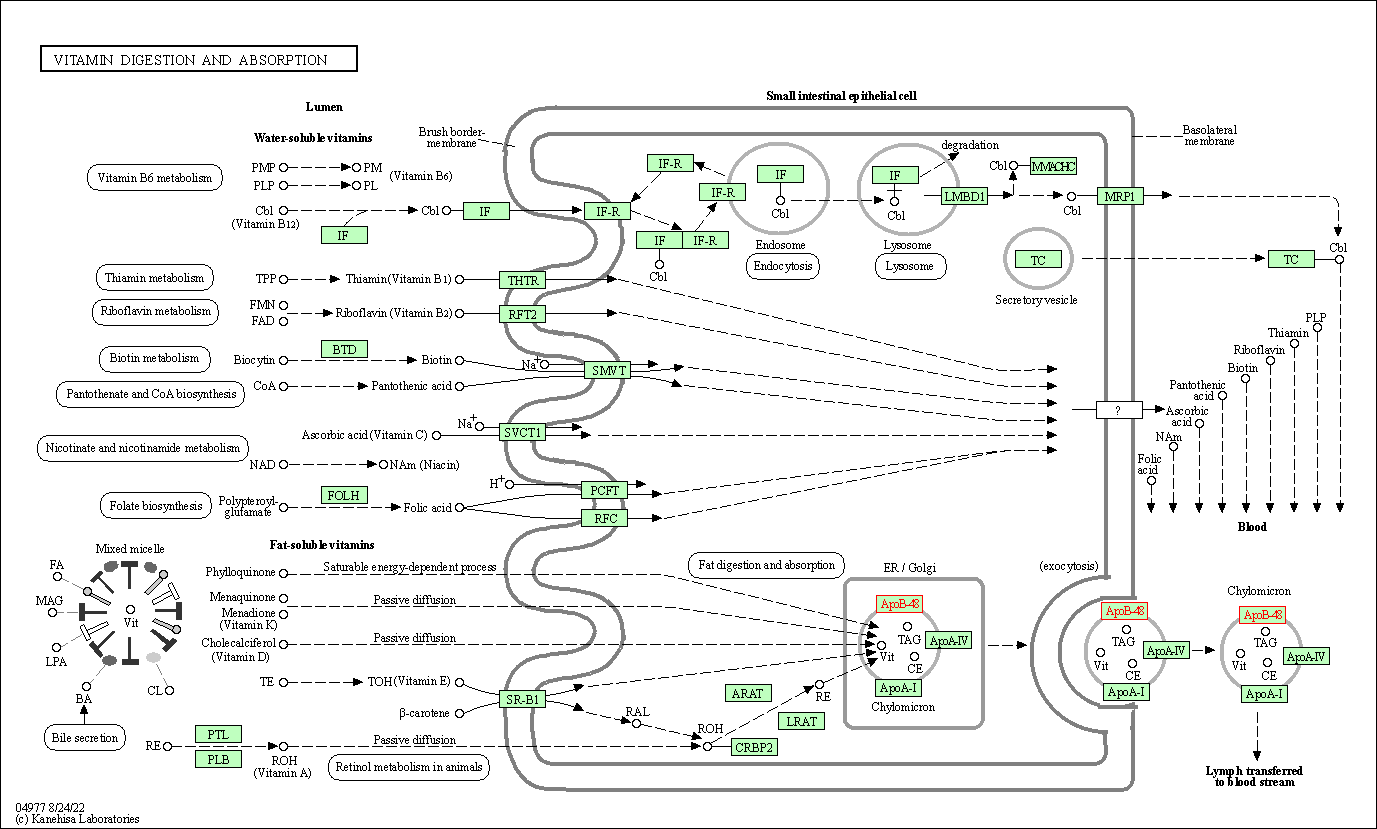
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Cholesterol metabolism | hsa04979 | Affiliated Target |
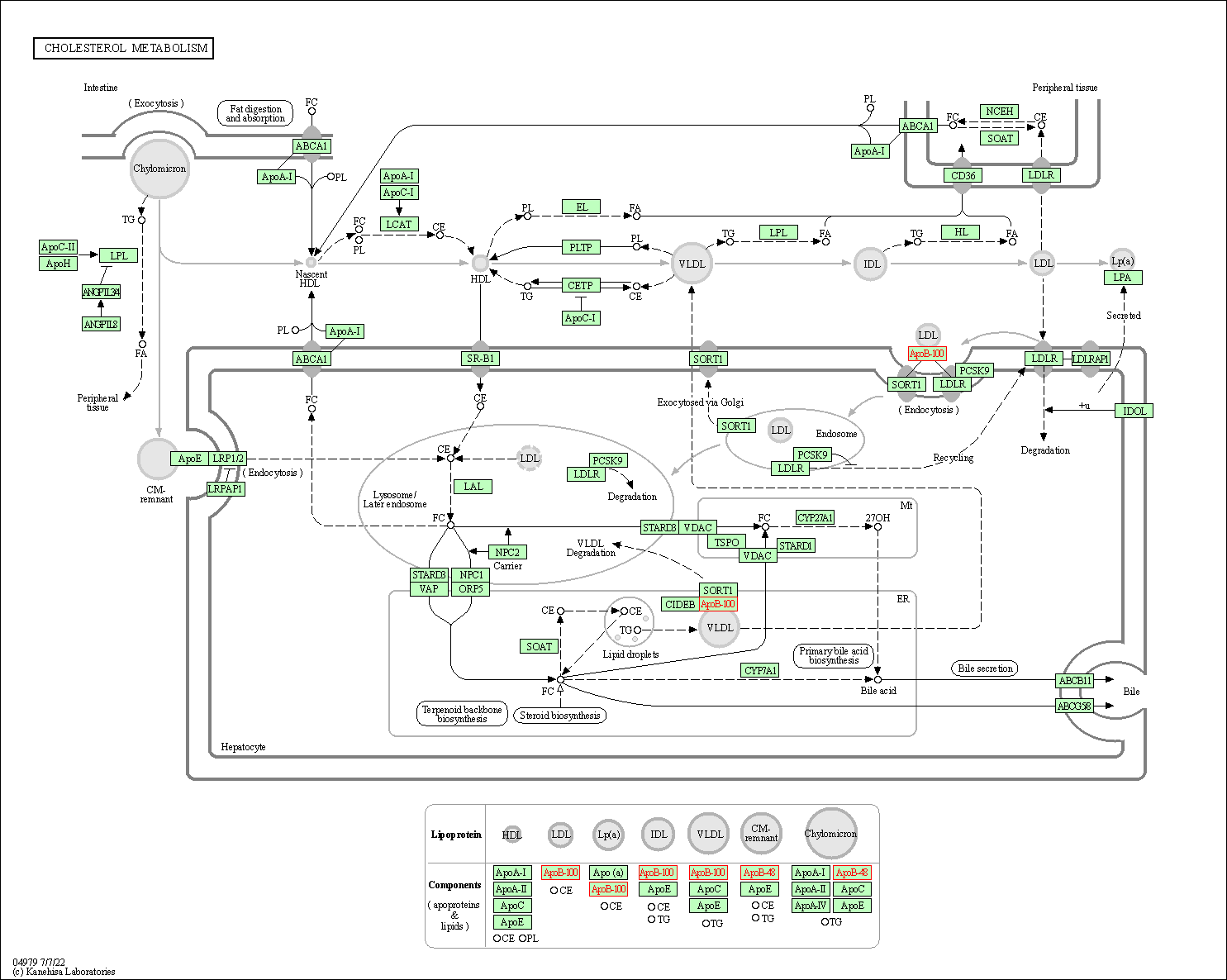
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 24 | Degree centrality | 2.58E-03 | Betweenness centrality | 1.77E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.07E-01 | Radiality | 1.36E+01 | Clustering coefficient | 1.92E-01 |
| Neighborhood connectivity | 1.21E+01 | Topological coefficient | 8.54E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 2 KEGG Pathways | + | ||||
| 1 | Fat digestion and absorption | |||||
| 2 | Vitamin digestion and absorption | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | amb2 Integrin signaling | |||||
| 2 | FOXA1 transcription factor network | |||||
| Reactome | [+] 8 Reactome Pathways | + | ||||
| 1 | LDL-mediated lipid transport | |||||
| 2 | Chylomicron-mediated lipid transport | |||||
| 3 | Cell surface interactions at the vascular wall | |||||
| 4 | Scavenging by Class B Receptors | |||||
| 5 | Scavenging by Class A Receptors | |||||
| 6 | Scavenging by Class H Receptors | |||||
| 7 | Platelet sensitization by LDL | |||||
| 8 | Retinoid metabolism and transport | |||||
| WikiPathways | [+] 9 WikiPathways | + | ||||
| 1 | Statin Pathway | |||||
| 2 | Binding and Uptake of Ligands by Scavenger Receptors | |||||
| 3 | Visual phototransduction | |||||
| 4 | Lipid digestion, mobilization, and transport | |||||
| 5 | Platelet homeostasis | |||||
| 6 | Cell surface interactions at the vascular wall | |||||
| 7 | Folate Metabolism | |||||
| 8 | Vitamin B12 Metabolism | |||||
| 9 | Selenium Micronutrient Network | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | 2011 Pipeline of Santaris Pharma. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

