Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T09423
(Former ID: TTDS00431)
|
|||||
| Target Name |
Inward rectifier potassium channel Kir1.2 (KCNJ10)
|
|||||
| Synonyms |
Potassiumchannel, inwardly rectifying, subfamily J, member 10; Potassium channel, inwardly rectifying subfamily J member 10; Inward rectifier K+ channel Kir1.2; Inward rectifier K(+) channel Kir1.2; ATP-sensitive inward rectifier potassium channel 10; ATP-dependent inwardly rectifying potassium channel Kir4.1; ATP dependent K+ channel
Click to Show/Hide
|
|||||
| Gene Name |
KCNJ10
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Acute diabete complication [ICD-11: 5A2Y] | |||||
| 2 | Type 2 diabetes mellitus [ICD-11: 5A11] | |||||
| Function |
Inward rectifier potassium channels are characterized by a greater tendency to allow potassium to flow into the cell rather than out of it. Their voltage dependence is regulated by the concentration of extracellular potassium; as external potassium is raised, the voltage range of the channel opening shifts to more positive voltages. The inward rectification is mainly due to the blockage of outward current by internal magnesium. Can be blocked by extracellular barium and cesium. In the kidney, together with KCNJ16, mediates basolateral K(+) recycling in distal tubules; this process is critical for Na(+) reabsorption at the tubules. May be responsible for potassium buffering action of glial cells in the brain.
Click to Show/Hide
|
|||||
| BioChemical Class |
Inward rectifier potassium channel
|
|||||
| UniProt ID | ||||||
| Sequence |
MTSVAKVYYSQTTQTESRPLMGPGIRRRRVLTKDGRSNVRMEHIADKRFLYLKDLWTTFI
DMQWRYKLLLFSATFAGTWFLFGVVWYLVAVAHGDLLELDPPANHTPCVVQVHTLTGAFL FSLESQTTIGYGFRYISEECPLAIVLLIAQLVLTTILEIFITGTFLAKIARPKKRAETIR FSQHAVVASHNGKPCLMIRVANMRKSLLIGCQVTGKLLQTHQTKEGENIRLNQVNVTFQV DTASDSPFLILPLTFYHVVDETSPLKDLPLRSGEGDFELVLILSGTVESTSATCQVRTSY LPEEILWGYEFTPAISLSASGKYIADFSLFDQVVKVASPSGLRDSTVRYGDPEKLKLEES LREQAEKEGSALSVRISNV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 6 Approved Drugs | + | ||||
| 1 | Chlorpropamide | Drug Info | Approved | Non-insulin dependent diabetes | [4], [5], [6] | |
| 2 | Gliquidone | Drug Info | Approved | Diabetic complication | [7] | |
| 3 | Glisoxepide | Drug Info | Approved | Non-insulin dependent diabetes | [8], [6] | |
| 4 | Glycodiazine | Drug Info | Approved | Diabetic complication | [9] | |
| 5 | Mitiglinide | Drug Info | Approved | Diabetic complication | [10] | |
| 6 | Tolazamide | Drug Info | Approved | Diabetic complication | [11], [12] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Blocker | [+] 5 Blocker drugs | + | ||||
| 1 | Chlorpropamide | Drug Info | [13] | |||
| 2 | Glisoxepide | Drug Info | [8] | |||
| 3 | Mitiglinide | Drug Info | [1] | |||
| 4 | Tolazamide | Drug Info | [15] | |||
| 5 | Sulfonylthioureas | Drug Info | [16] | |||
| Binder | [+] 2 Binder drugs | + | ||||
| 1 | Gliquidone | Drug Info | [14] | |||
| 2 | Glycodiazine | Drug Info | [9] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Gastric acid secretion | hsa04971 | Affiliated Target |
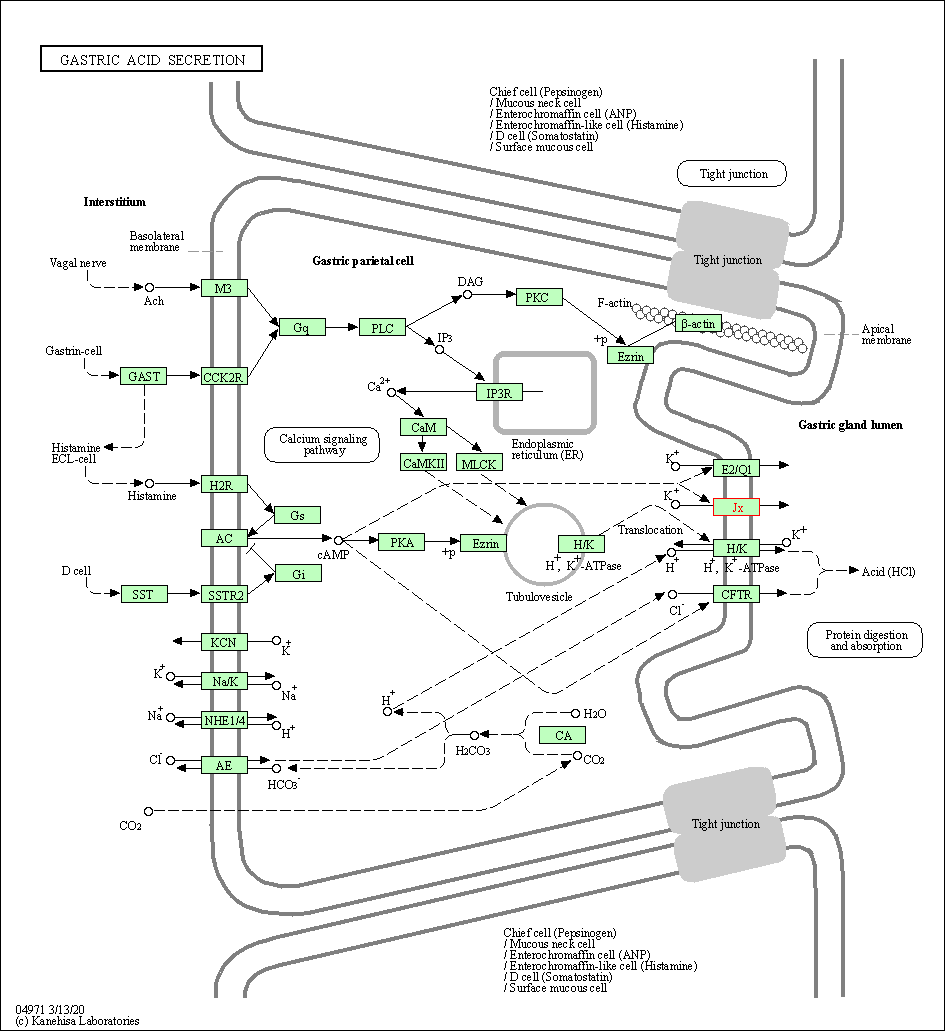
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.56E-01 | Radiality | 1.22E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 5.00E+00 | Topological coefficient | 1.00E+00 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Gastric acid secretion | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Activation of G protein gated Potassium channels | |||||
| 2 | Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | |||||
| 2 | Potassium Channels | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target QSAR Model | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Carboxyl-glucuronidation of mitiglinide by human UDP-glucuronosyltransferases. Biochem Pharmacol. 2007 Jun 1;73(11):1842-51. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6821). | |||||
| REF 3 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (ANDA) 074223. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6801). | |||||
| REF 5 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (ANDA) 088548. | |||||
| REF 6 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 7 | Drug information of Gliquidone, 2008. eduDrugs. | |||||
| REF 8 | Bepridil, an antiarrhythmic drug, opens mitochondrial KATP channels, blocks sarcolemmal KATP channels, and confers cardioprotection. J Pharmacol Exp Ther. 2006 Jan;316(1):182-8. | |||||
| REF 9 | Coupling between proximal tubular transport processes. Studies with ouabain, SITS and HCO3-free solutions. Pflugers Arch. 1977 Apr 25;368(3):245-52. | |||||
| REF 10 | Drug information of Mitiglinide, 2008. eduDrugs. | |||||
| REF 11 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6847). | |||||
| REF 12 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 018894. | |||||
| REF 13 | Effect of the chlorpropamide and fructose-1,6-bisphosphate of soluble TNF receptor II levels. Pharmacol Res. 2004 May;49(5):449-53. | |||||
| REF 14 | Hydroxyl-substituted sulfonylureas as potent inhibitors of specific [3H]glyburide binding to rat brain synaptosomes. Bioorg Med Chem. 2003 May 1;11(9):2099-113. | |||||
| REF 15 | Inhibition of ATP-activated potassium channels exerts pressor effects and improves survival in a rat model of severe hemorrhagic shock. Shock. 1996 Jun;5(6):391-4. | |||||
| REF 16 | Cardioselective K(ATP) channel blockers derived from a new series of m-anisamidoethylbenzenesulfonylthioureas. J Med Chem. 2001 Mar 29;44(7):1085-98. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

