Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T10735
(Former ID: TTDC00027)
|
|||||
| Target Name |
Histidine decarboxylase (HDC)
|
|||||
| Synonyms |
Human histidine decarboxylase
Click to Show/Hide
|
|||||
| Gene Name |
HDC
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Hypo-osmolality/hyponatraemia [ICD-11: 5C72] | |||||
| Function |
Catalyzes the biosynthesis of histamine from histidine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Carbon-carbon lyase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 4.1.1.22
|
|||||
| Sequence |
MMEPEEYRERGREMVDYICQYLSTVRERRVTPDVQPGYLRAQLPESAPEDPDSWDSIFGD
IERIIMPGVVHWQSPHMHAYYPALTSWPSLLGDMLADAINCLGFTWASSPACTELEMNVM DWLAKMLGLPEHFLHHHPSSQGGGVLQSTVSESTLIALLAARKNKILEMKTSEPDADESC LNARLVAYASDQAHSSVEKAGLISLVKMKFLPVDDNFSLRGEALQKAIEEDKQRGLVPVF VCATLGTTGVCAFDCLSELGPICAREGLWLHIDAAYAGTAFLCPEFRGFLKGIEYADSFT FNPSKWMMVHFDCTGFWVKDKYKLQQTFSVNPIYLRHANSGVATDFMHWQIPLSRRFRSV KLWFVIRSFGVKNLQAHVRHGTEMAKYFESLVRNDPSFEIPAKRHLGLVVFRLKGPNCLT ENVLKEIAKAGRLFLIPATIQDKLIIRFTVTSQFTTRDDILRDWNLIRDAATLILSQHCT SQPSPRVGNLISQIRGARAWACGTSLQSVSGAGDDPVQARKIIKQPQRVGAGPMKRENGL HLETLLDPVDDCFSEEAPDATKHKLSSFLFSYLSVQTKKKTVRSLSCNSVPVSAQKPLPT EASVKNGGSSRVRIFSRFPEDMMMLKKSAFKKLIKFYSVPSFPECSSQCGLQLPCCPLQA MV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T76AZQ | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Lixivaptan | Drug Info | Phase 3 | Hyponatraemia | [2], [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 3 Inhibitor drugs | + | ||||
| 1 | Lixivaptan | Drug Info | [1] | |||
| 2 | AMA | Drug Info | [5] | |||
| 3 | Brocresine | Drug Info | [6], [7] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Pyridoxal phosphate | Ligand Info | |||||
| Structure Description | Human histidine decarboxylase complex with Histidine methyl ester (HME) | PDB:4E1O | ||||
| Method | X-ray diffraction | Resolution | 1.80 Å | Mutation | Yes | [8] |
| PDB Sequence |
SMEPEEYRER
10 GREMVDYICQ20 YLSTVRERRV30 TPDVQPGYLR40 AQLPESAPED50 PDSWDSIFGD 60 IERIIMPGVV70 HWQSPHMHAY80 YPALTSWPSL90 LGDMLADAIN100 CLGFTWASSP 110 ACTELEMNVM120 DWLAKMLGLP130 EHFLHHHPSS140 QGGGVLQSTV150 SESTLIALLA 160 ARKNKILEMK170 TSEPDADESS180 LNARLVAYAS190 DQAHSSVEKA200 GLISLVKMKF 210 LPVDDNFSLR220 GEALQKAIEE230 DKQRGLVPVF240 VCATLGTTGV250 CAFDLSELGP 261 ICAREGLWLH271 IDAAYAGTAF281 LCPEFRGFLK291 GIEYADSFTF301 NPSKWMMVHF 311 DCTGFWVKDK321 YKLQQTFSVN331 PIYLRHANSG341 VATDFMHWQI351 PLSRRFRSVK 361 LWFVIRSFGV371 KNLQAHVRHG381 TEMAKYFESL391 VRNDPSFEIP401 AKRHLGLVVF 411 RLKGPNSLTE421 NVLKEIAKAG431 RLFLIPATIQ441 DKLIIRFTVT451 SQFTTRDDIL 461 RDWNLIRDAA471 TLILSQ
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Cysteine Sulfenic Acid | Ligand Info | |||||
| Structure Description | Human histidine decarboxylase complex with Histidine methyl ester (HME) | PDB:4E1O | ||||
| Method | X-ray diffraction | Resolution | 1.80 Å | Mutation | Yes | [8] |
| PDB Sequence |
SMEPEEYRER
10 GREMVDYICQ20 YLSTVRERRV30 TPDVQPGYLR40 AQLPESAPED50 PDSWDSIFGD 60 IERIIMPGVV70 HWQSPHMHAY80 YPALTSWPSL90 LGDMLADAIN100 CLGFTWASSP 110 ACTELEMNVM120 DWLAKMLGLP130 EHFLHHHPSS140 QGGGVLQSTV150 SESTLIALLA 160 ARKNKILEMK170 TSEPDADESS180 LNARLVAYAS190 DQAHSSVEKA200 GLISLVKMKF 210 LPVDDNFSLR220 GEALQKAIEE230 DKQRGLVPVF240 VCATLGTTGV250 CAFDLSELGP 261 ICAREGLWLH271 IDAAYAGTAF281 LCPEFRGFLK291 GIEYADSFTF301 NPSKWMMVHF 311 DCTGFWVKDK321 YKLQQTFSVN331 PIYLRHANSG341 VATDFMHWQI351 PLSRRFRSVK 361 LWFVIRSFGV371 KNLQAHVRHG381 TEMAKYFESL391 VRNDPSFEIP401 AKRHLGLVVF 411 RLKGPNSLTE421 NVLKEIAKAG431 RLFLIPATIQ441 DKLIIRFTVT451 SQFTTRDDIL 461 RDWNLIRDAA471 TLILSQ
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Histidine metabolism | hsa00340 | Affiliated Target |
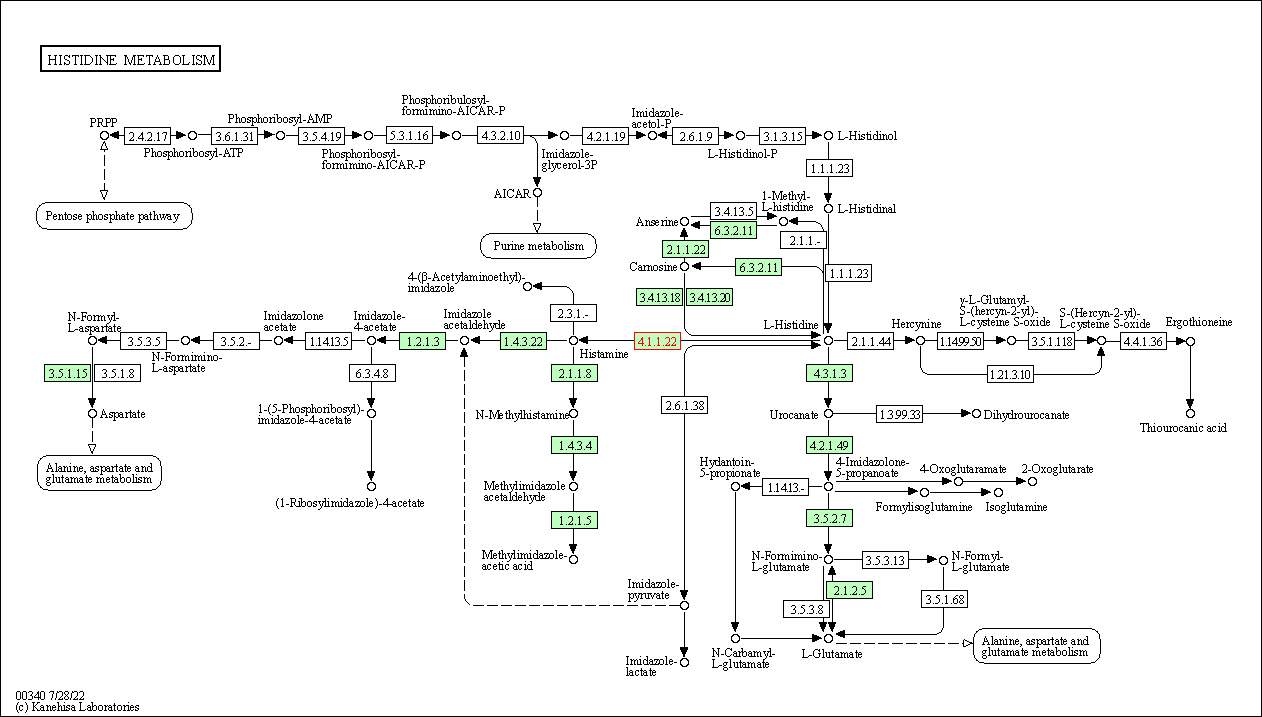
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.61E-01 | Radiality | 1.24E+01 | Clustering coefficient | 1.00E+00 |
| Neighborhood connectivity | 4.00E+00 | Topological coefficient | 6.67E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating Transcription Factors | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 2 KEGG Pathways | + | ||||
| 1 | Histidine metabolism | |||||
| 2 | Metabolic pathways | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Histamine synthesis | |||||
| 2 | CCKR signaling map ST | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Histidine Metabolism | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | Biogenic Amine Synthesis | |||||
| 2 | Metabolism of amino acids and derivatives | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of Biofrontera (2009). | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 2238). | |||||
| REF 3 | Physical state of kappa-carrageenan modulates the mode of action of kappa-carrageenase from Pseudoalteromonas carrageenovora. Biochem J. 2009 May 1;419(3):545-53. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5189). | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1274). | |||||
| REF 6 | Histamine in brain--its role in regulation of seizure susceptibility. Epilepsy Res. 1991 Nov-Dec;10(2-3):111-8. | |||||
| REF 7 | Effects of brocresine (NSD-1055) and cycloheximide on amino acid decarboxylase activities in gastric mucosa of normal and vagally denervated rats. Br J Pharmacol. 1972 Dec;46(4):688-95. | |||||
| REF 8 | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase. J Biol Chem. 2012 Aug 17;287(34):29175-83. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

