Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T12817
(Former ID: TTDS00234)
|
|||||
| Target Name |
Lipopolysaccharide-associated protein 1 (HSPA8)
|
|||||
| Synonyms |
LPS-associated protein 1; LAP-1; Heat shock protein 73; Heat shock cognate 71 kDa protein; Heat shock 70 kDa protein 8; HSPA10; HSP73; HSC70
Click to Show/Hide
|
|||||
| Gene Name |
HSPA8
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Plays a pivotal role in the protein quality control system, ensuring the correct folding of proteins, the re-folding of misfolded proteins and controlling the targeting of proteins for subsequent degradation. This is achieved through cycles of ATP binding, ATP hydrolysis and ADP release, mediated by co-chaperones. The co-chaperones have been shown to not only regulate different steps of the ATPase cycle of HSP70, but they also have an individual specificity such that one co-chaperone may promote folding of a substrate while another may promote degradation. The affinity of HSP70 for polypeptides is regulated by its nucleotide bound state. In the ATP-bound form, it has a low affinity for substrate proteins. However, upon hydrolysis of the ATP to ADP, it undergoes a conformational change that increases its affinity for substrate proteins. HSP70 goes through repeated cycles of ATP hydrolysis and nucleotide exchange, which permits cycles of substrate binding and release. The HSP70-associated co-chaperones are of three types: J-domain co-chaperones HSP40s (stimulate ATPase hydrolysis by HSP70), the nucleotide exchange factors (NEF) such as BAG1/2/3 (facilitate conversion of HSP70 from the ADP-bound to the ATP-bound state thereby promoting substrate release), and the TPR domain chaperones such as HOPX and STUB1. Acts as a repressor of transcriptional activation. Inhibits the transcriptional coactivator activity of CITED1 on Smad-mediated transcription. Component of the PRP19-CDC5L complex that forms an integral part of the spliceosome and is required for activating pre-mRNA splicing. May have a scaffolding role in the spliceosome assembly as it contacts all other components of the core complex. Binds bacterial lipopolysaccharide (LPS) and mediates LPS-induced inflammatory response, including TNF secretion by monocytes. Participates in the ER-associated degradation (ERAD) quality control pathway in conjunction with J domain-containing co-chaperones and the E3 ligase STUB1. Interacts with VGF-derived peptide TLQP-21. Molecular chaperone implicated in a wide variety of cellular processes, including protection of the proteome from stress, folding and transport of newly synthesized polypeptides, activation of proteolysis of misfolded proteins and the formation and dissociation of protein complexes.
Click to Show/Hide
|
|||||
| BioChemical Class |
Heat shock protein
|
|||||
| UniProt ID | ||||||
| Sequence |
MSKGPAVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVA
MNPTNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVNDAGRPKVQVEYKGETKSFYPEEVS SMVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAA IAYGLDKKVGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDFDNRMVNH FIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEIDSLYEGIDFYTSITRA RFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELN KSINPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTI PTKQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDI DANGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKN SLESYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIINWLDKNQTAEKEEFEHQQKELE KVCNPIITKLYQSAGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine | Ligand Info | |||||
| Structure Description | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | PDB:5AQF | ||||
| Method | X-ray diffraction | Resolution | 1.88 Å | Mutation | No | [3] |
| PDB Sequence |
SMSKGPAVGI
9 DLGTTYSCVG19 VFQHGKVEII29 ANDQGNRTTP39 SYVAFTDTER49 LIGDAAKNQV 59 AMNPTNTVFD69 AKRLIGRRFD79 DAVVQSDMKH89 WPFMVVNDAG99 RPKVQVEYKG 109 ETKSFYPEEV119 SSMVLTKMKE129 IAEAYLGKTV139 TNAVVTVPAY149 FNDSQRQATK 159 DAGTIAGLNV169 LRIINEPTAA179 AIAYGLDKKV189 GAERNVLIFD199 LGGGTFDVSI 209 LTIEDGIFEV219 KSTAGDTHLG229 GEDFDNRMVN239 HFIAEFKRKH249 KKDISENKRA 259 VRRLRTACER269 AKRTLSSSTQ279 ASIEIDSLYE289 GIDFYTSITR299 ARFEELNADL 309 FRGTLDPVEK319 ALRDAKLDKS329 QIHDIVLVGG339 STRIPKIQKL349 LQDFFNGKEL 359 NKSINPDEAV369 AYGAAVQAAI379 LS
|
|||||
|
|
||||||
| Ligand Name: Adenine | Ligand Info | |||||
| Structure Description | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | PDB:5AQI | ||||
| Method | X-ray diffraction | Resolution | 1.98 Å | Mutation | No | [3] |
| PDB Sequence |
MSKGPAVGID
10 LGTTYSCVGV20 FQHGKVEIIA30 NDQGNRTTPS40 YVAFTDTERL50 IGDAAKNQVA 60 MNPTNTVFDA70 KRLIGRRFDD80 AVVQSDMKHW90 PFMVVNDAGR100 PKVQVEYKGE 110 TKSFYPEEVS120 SMVLTKMKEI130 AEAYLGKTVT140 NAVVTVPAYF150 NDSQRQATKD 160 AGTIAGLNVL170 RIINEPTAAA180 IAYGLDKKVG190 AERNVLIFDL200 GGGTFDVSIL 210 TIEDGIFEVK220 STAGDTHLGG230 EDFDNRMVNH240 FIAEFKRKHK250 KDISENKRAV 260 RRLRTACERA270 KRTLSSSTQA280 SIEIDSLYEG290 IDFYTSITRA300 RFEELNADLF 310 RGTLDPVEKA320 LRDAKLDKSQ330 IHDIVLVGGS340 TRIPKIQKLL350 QDFFNGKELN 360 KSINPDEAVA370 YGAAVQAAIL380 S
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Ankyrin repeat domain-containing protein 45 (ANKRD45) | 33.333 (26/78) | 2.99E-04 |
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Spliceosome | hsa03040 | Affiliated Target |

|
| Class: Genetic Information Processing => Transcription | Pathway Hierarchy | ||
| MAPK signaling pathway | hsa04010 | Affiliated Target |
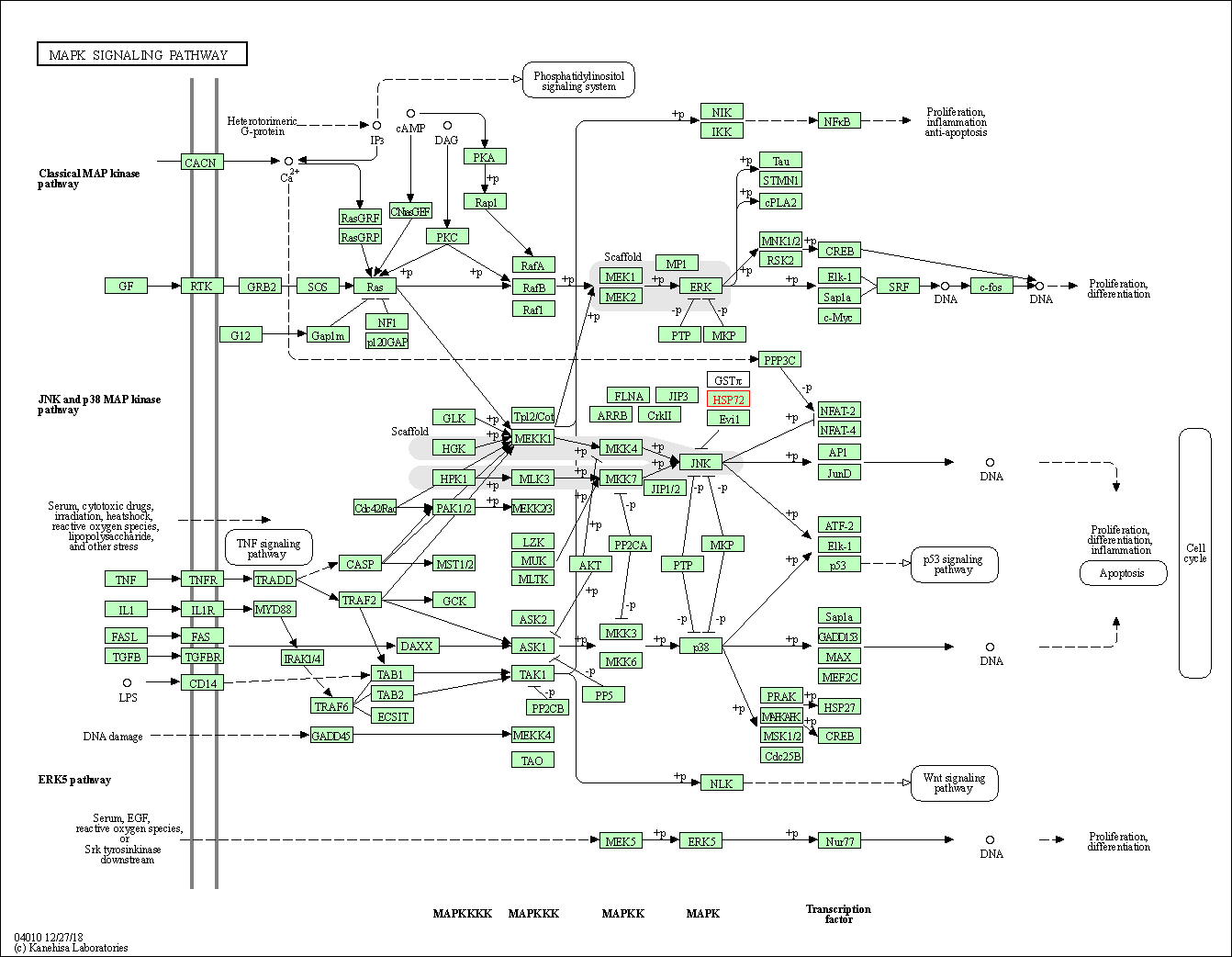
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |
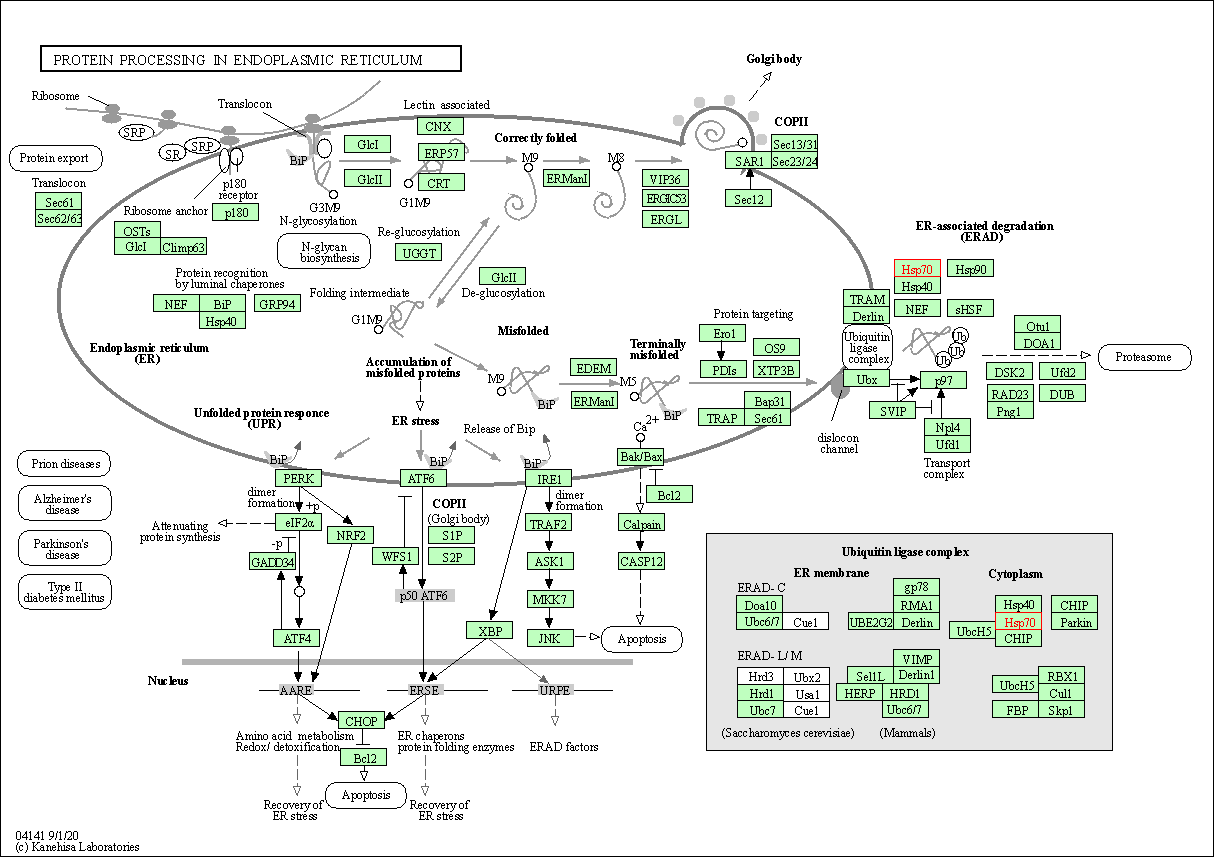
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Antigen processing and presentation | hsa04612 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 59 | Degree centrality | 6.34E-03 | Betweenness centrality | 1.73E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.64E-01 | Radiality | 1.45E+01 | Clustering coefficient | 5.32E-02 |
| Neighborhood connectivity | 2.62E+01 | Topological coefficient | 2.81E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 2 | In silico identification and biochemical evaluation of novel inhibitors of NRH:quinone oxidoreductase 2 (NQO2). Bioorg Med Chem Lett. 2010 Dec 15;20(24):7331-6. | |||||
| REF 3 | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms. Sci Rep. 2016 Oct 6;6:34701. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

