Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T13176
(Former ID: TTDI03189)
|
|||||
| Target Name |
PRKR-like endoplasmic reticulum kinase (PERK)
|
|||||
| Synonyms |
PEK
Click to Show/Hide
|
|||||
| Gene Name |
EIF2AK3
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Breast cancer [ICD-11: 2C60-2C6Y] | |||||
| Function |
Converts phosphorylated eIF-2-alpha/EIF2S1 either in a global protein synthesis inhibitor, leading to a reduced overall utilization of amino acids, or to a translation initiation activator of specific mRNAs, such as the transcriptional activator ATF4, and hence allowing ATF4-mediated reprogramming of amino acid biosynthetic gene expression to alleviate nutrient depletion. Serves as a critical effector of unfolded protein response (UPR)-induced G1 growth arrest due to the loss of cyclin-D1 (CCND1). Involved in control of mitochondrial morphology and function. Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2-alpha/EIF2S1) on 'Ser-52' during the unfolded protein response (UPR) and in response to low amino acid availability.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MERAISPGLLVRALLLLLLLLGLAARTVAAGRARGLPAPTAEAAFGLGAAAAPTSATRVP
AAGAVAAAEVTVEDAEALPAAAGEQEPRGPEPDDETELRPRGRSLVIISTLDGRIAALDP ENHGKKQWDLDVGSGSLVSSSLSKPEVFGNKMIIPSLDGALFQWDQDRESMETVPFTVES LLESSYKFGDDVVLVGGKSLTTYGLSAYSGKVRYICSALGCRQWDSDEMEQEEDILLLQR TQKTVRAVGPRSGNEKWNFSVGHFELRYIPDMETRAGFIESTFKPNENTEESKIISDVEE QEAAIMDIVIKVSVADWKVMAFSKKGGHLEWEYQFCTPIASAWLLKDGKVIPISLFDDTS YTSNDDVLEDEEDIVEAARGATENSVYLGMYRGQLYLQSSVRISEKFPSSPKALESVTNE NAIIPLPTIKWKPLIHSPSRTPVLVGSDEFDKCLSNDKFSHEEYSNGALSILQYPYDNGY YLPYYKRERNKRSTQITVRFLDNPHYNKNIRKKDPVLLLHWWKEIVATILFCIIATTFIV RRLFHPHPHRQRKESETQCQTENKYDSVSGEANDSSWNDIKNSGYISRYLTDFEPIQCLG RGGFGVVFEAKNKVDDCNYAIKRIRLPNRELAREKVMREVKALAKLEHPGIVRYFNAWLE APPEKWQEKMDEIWLKDESTDWPLSSPSPMDAPSVKIRRMDPFATKEHIEIIAPSPQRSR SFSVGISCDQTSSSESQFSPLEFSGMDHEDISESVDAAYNLQDSCLTDCDVEDGTMDGND EGHSFELCPSEASPYVRSRERTSSSIVFEDSGCDNASSKEEPKTNRLHIGNHCANKLTAF KPTSSKSSSEATLSISPPRPTTLSLDLTKNTTEKLQPSSPKVYLYIQMQLCRKENLKDWM NGRCTIEERERSVCLHIFLQIAEAVEFLHSKGLMHRDLKPSNIFFTMDDVVKVGDFGLVT AMDQDEEEQTVLTPMPAYARHTGQVGTKLYMSPEQIHGNSYSHKVDIFSLGLILFELLYP FSTQMERVRTLTDVRNLKFPPLFTQKYPCEYVMVQDMLSPSPMERPEAINIIENAVFEDL DFPGKTVLRQRSRSLSSSGTKHSRQSNNSHSPLPSN Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T22V6A | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: GSK2606414 | Ligand Info | |||||
| Structure Description | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | PDB:4G31 | ||||
| Method | X-ray diffraction | Resolution | 2.28 Å | Mutation | No | [3] |
| PDB Sequence |
GRYLTDFEPI
595 QCLGRGGVVF607 EAKNKVDDCN617 YAIKRIRLPN627 RELAREKVMR637 EVKALAKLEH 647 PGIVRYFNAW657 LEKNKVYLYI885 QMQLCRKENL895 KDWMNGRCTI905 EERERSVCLH 915 IFLQIAEAVE925 FLHSKGLMHR935 DLKPSNIFFT945 MDDVVKVGDF955 GLVGTKLYMS 991 PEQIHGNSYS1001 HKVDIFSLGL1011 ILFELLYPFS1021 TQMERVRTLT1031 DVRNLKFPPL 1041 FTQKYPCEYV1051 MVQDMLSPSP1061 MERPEAINII1071 ENAVFEDL
|
|||||
|
|
LEU598
3.817
GLY599
4.990
VAL606
3.923
ALA619
3.389
LYS621
3.877
GLU638
4.609
VAL639
3.402
LEU642
3.534
ALA643
3.169
ILE650
4.436
VAL651
3.637
TYR653
3.393
ALA656
4.583
|
|||||
| Ligand Name: 4-{2-Amino-3-[5-Fluoro-2-(Methylamino)quinazolin-6-Yl]-4-Methylbenzoyl}-1-Methyl-2,5-Diphenyl-1,2-Dihydro-3h-Pyrazol-3-One | Ligand Info | |||||
| Structure Description | Co-crystal Structure of PERK bound to 4-{2-amino-3-[5-fluoro-2-(methylamino)quinazolin-6-yl]-4-methylbenzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | PDB:4X7K | ||||
| Method | X-ray diffraction | Resolution | 1.80 Å | Mutation | Yes | [4] |
| PDB Sequence |
YISRYLTDFE
594 PIQCLGRGGF604 GVVFEAKNKV614 DDCNYAIKRI624 RLPNRELARE634 KVMREVKALA 644 KLEHPGIVRY654 FNAWLEAPPK881 VYLYIQMQLC891 RKENLKDWMN901 GRCTIEERER 911 SVCLHIFLQI921 AEAVEFLHSK931 GLMHRNLKPS941 NIFFTMDDVV951 KVGDFGLVTT 987 KLYMSPEQIH997 GNSYSHKVDI1007 FSLGLILFEL1017 LYPFSTQMER1027 VRTLTDVRNL 1037 KFPPLFTQKY1047 PCEYVMVQDM1057 LSPSPMERPE1067 AINIIENAVF1077 EDLDFPGKTV 1087
|
|||||
|
|
LEU599
3.904
VAL607
3.675
ALA620
3.402
ILE621
4.021
LYS622
2.729
ILE624
3.790
VAL636
4.009
GLU639
3.208
VAL640
4.542
LEU643
3.522
LEU646
4.185
ILE651
3.797
VAL652
3.448
ILE886
3.663
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Mitophagy - animal | hsa04137 | Affiliated Target |
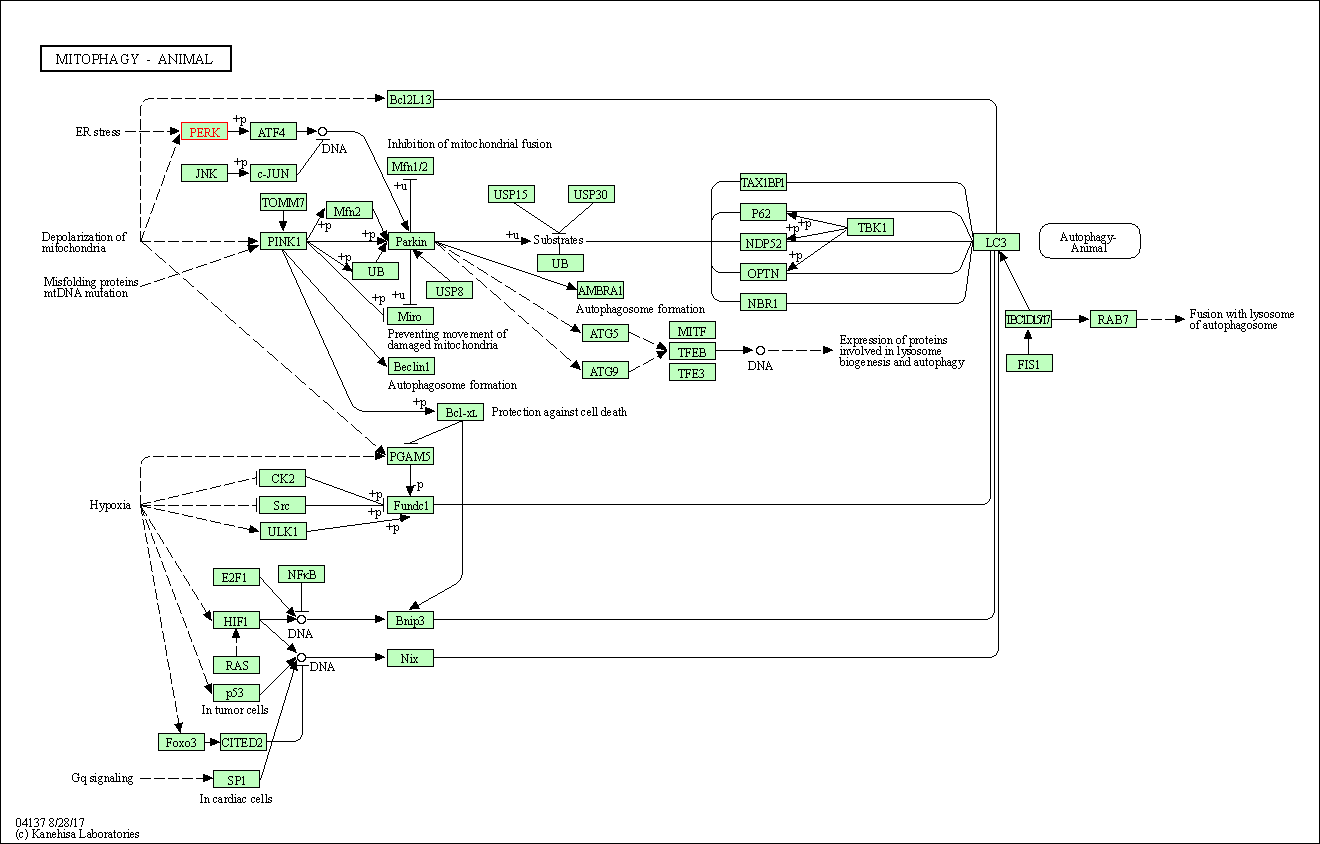
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
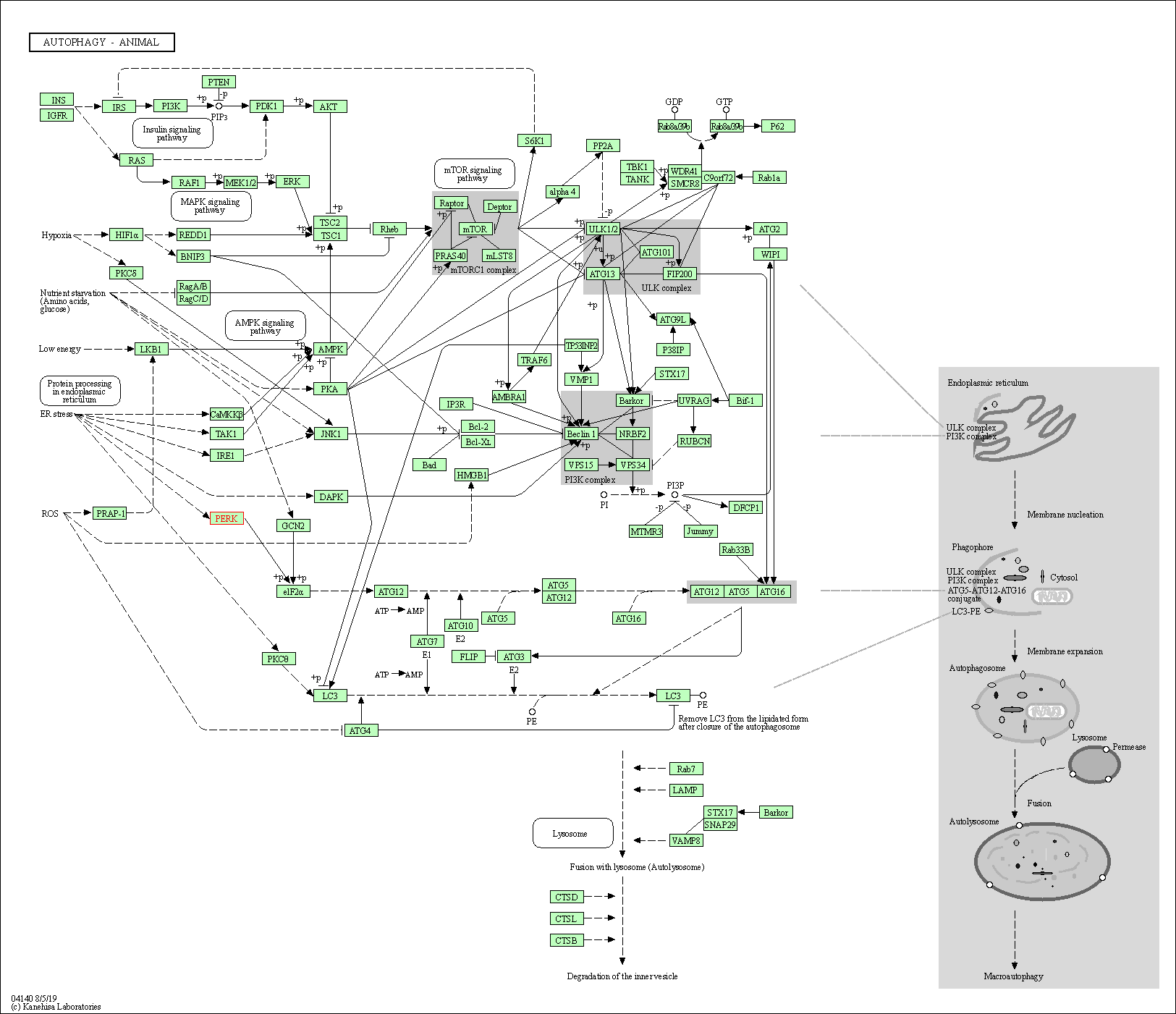
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |
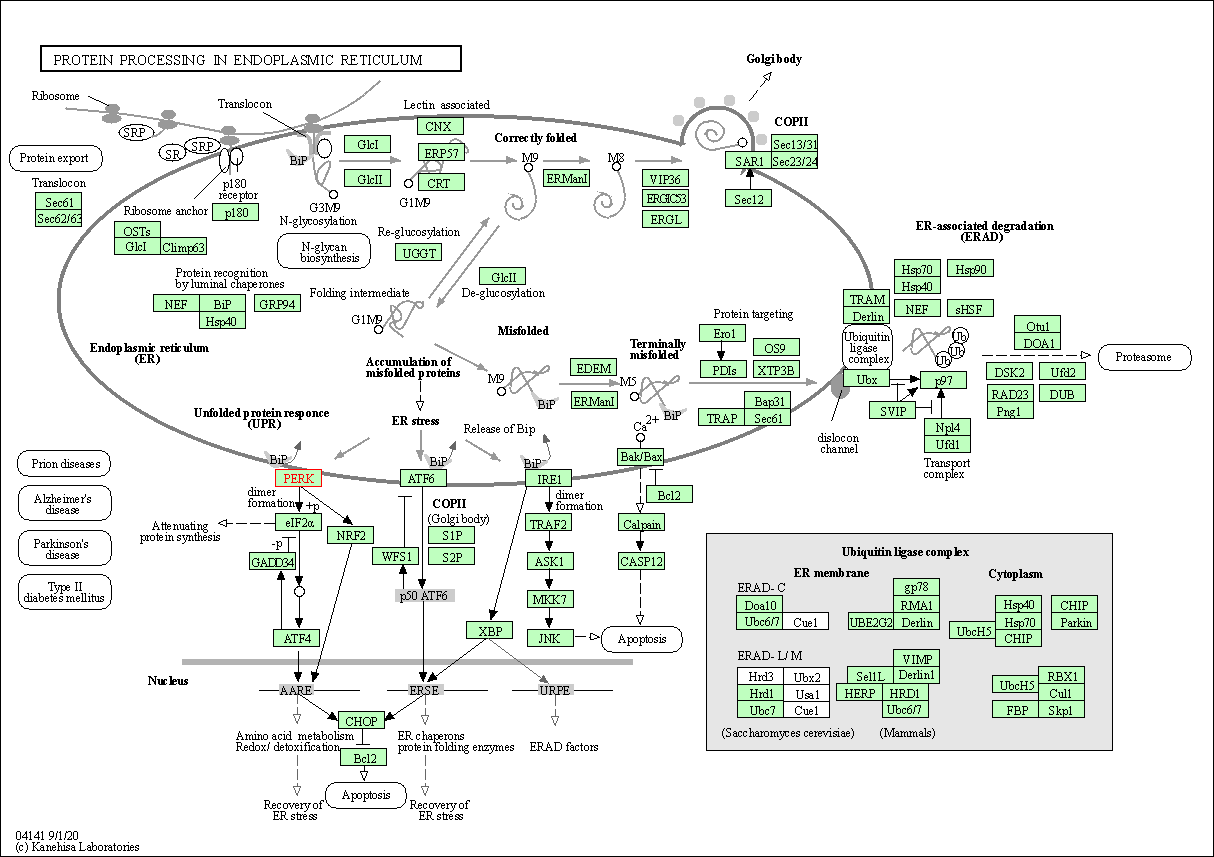
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 6.02E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.14E-01 | Radiality | 1.37E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 3.24E+01 | Topological coefficient | 2.12E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Protein kinase R(PKR)-like endoplasmic reticulum kinase (PERK) inhibitors: a patent review (2010-2015).Expert Opin Ther Pat. 2017 Jan;27(1):37-48. | |||||
| REF 2 | ClinicalTrials.gov (NCT04834778) A Multicenter, Open-label, Phase 1a Study of HC-5404-FU in Subjects With Advanced Solid Tumors. U.S.National Institutes of Health. | |||||
| REF 3 | Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem. 2012 Aug 23;55(16):7193-207. | |||||
| REF 4 | Discovery of 1H-pyrazol-3(2H)-ones as potent and selective inhibitors of protein kinase R-like endoplasmic reticulum kinase (PERK). J Med Chem. 2015 Feb 12;58(3):1426-41. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

