Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T14110
(Former ID: TTDNR00650)
|
|||||
| Target Name |
Beta-klotho (KLB)
|
|||||
| Synonyms |
Klotho beta-like protein; BetaKlotho; BKL
Click to Show/Hide
|
|||||
| Gene Name |
KLB
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||||
| 2 | Type 2 diabetes mellitus [ICD-11: 5A11] | |||||
| Function |
Probably inactive as a glycosidase. Increases the ability of FGFR1 and FGFR4 to bind FGF21. Contributes to the transcriptional repression of cholesterol 7-alpha-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid synthesis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Glycosylase
|
|||||
| UniProt ID | ||||||
| Sequence |
MKPGCAAGSPGNEWIFFSTDEITTRYRNTMSNGGLQRSVILSALILLRAVTGFSGDGRAI
WSKNPNFTPVNESQLFLYDTFPKNFFWGIGTGALQVEGSWKKDGKGPSIWDHFIHTHLKN VSSTNGSSDSYIFLEKDLSALDFIGVSFYQFSISWPRLFPDGIVTVANAKGLQYYSTLLD ALVLRNIEPIVTLYHWDLPLALQEKYGGWKNDTIIDIFNDYATYCFQMFGDRVKYWITIH NPYLVAWHGYGTGMHAPGEKGNLAAVYTVGHNLIKAHSKVWHNYNTHFRPHQKGWLSITL GSHWIEPNRSENTMDIFKCQQSMVSVLGWFANPIHGDGDYPEGMRKKLFSVLPIFSEAEK HEMRGTADFFAFSFGPNNFKPLNTMAKMGQNVSLNLREALNWIKLEYNNPRILIAENGWF TDSRVKTEDTTAIYMMKNFLSQVLQAIRLDEIRVFGYTAWSLLDGFEWQDAYTIRRGLFY VDFNSKQKERKPKSSAHYYKQIIRENGFSLKESTPDVQGQFPCDFSWGVTESVLKPESVA SSPQFSDPHLYVWNATGNRLLHRVEGVRLKTRPAQCTDFVNIKKQLEMLARMKVTHYRFA LDWASVLPTGNLSAVNRQALRYYRCVVSEGLKLGISAMVTLYYPTHAHLGLPEPLLHADG WLNPSTAEAFQAYAGLCFQELGDLVKLWITINEPNRLSDIYNRSGNDTYGAAHNLLVAHA LAWRLYDRQFRPSQRGAVSLSLHADWAEPANPYADSHWRAAERFLQFEIAWFAEPLFKTG DYPAAMREYIASKHRRGLSSSALPRLTEAERRLLKGTVDFCALNHFTTRFVMHEQLAGSR YDSDRDIQFLQDITRLSSPTRLAVIPWGVRKLLRWVRRNYGDMDIYITASGIDDQALEDD RLRKYYLGKYLQEVLKAYLIDKVRIKGYYAFKLAEEKSKPRFGFFTSDFKAKSSIQFYNK VISSRGFPFENSSSRCSQTQENTECTVCLFLVQKKPLIFLGCCFFSTLVLLLSIAIFQRQ KRRKFWKAKNLQHIPLKKGKRVVS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | NGM313 | Drug Info | Phase 1 | Type-2 diabetes | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | NGM313 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Beta-D-Glucose | Ligand Info | |||||
| Structure Description | Crystal Structure of Beta-Klotho | PDB:5VAN | ||||
| Method | X-ray diffraction | Resolution | 2.20 Å | Mutation | Yes | [3] |
| PDB Sequence |
FSGDGRAIWS
62 QLFLYDTFPK83 NFFWGIGTGA93 LQVEGSWKKD103 GKGPSIWDHF113 IHTHLGSSDS 130 YIFLEKDLSA140 LDFIGVSFYQ150 FSISWPRLFP160 DGIVTVANAK170 GLQYYSTLLD 180 ALVLRNIEPI190 VTLYHWDLPL200 ALQEKYGGWK210 NDTIIDIFND220 YATYCFQMFG 230 DRVKYWITIH240 NPYLVAWHGY250 GTGMHAPGEK260 GNLAAVYTVG270 HNLIKAHSKV 280 WHNYNTHFRP290 HQKGWLSITL300 GSHWIEPQRS310 ENTMDIFKCQ320 QSMVSVLGWF 330 ANPIHGDGDY340 PEGMRKKLFS350 VLPIFSEAEK360 HEMRGTADFF370 AFSFGPNNFK 380 PLNTMAKMGQ390 NVSLNLREAL400 NWIKLEYNNP410 RILIAENGWF420 TDSRVKTEDT 430 TAIYMMKNFL440 SQVLQAIRLD450 EIRVFGYTAW460 SLLDGFEWQD470 AYTIRRGLFY 480 VDFNSKQKER490 KPKSSAHYYK500 QIIRENGFSL510 KESTPDVQGQ520 FPCDFSWGVT 530 ESVLKPEQCT577 DFVNIKKQLE587 MLARMKVTHY597 RFALDWASVL607 PTGQLSAVNR 617 QALRYYRCVV627 SEGLKLGISA637 MVTLYYPTHA647 HLGLPEPLLH657 ADGWLNPSTA 667 EAFQAYAGLC677 FQELGDLVKL687 WITINEPNRL697 SDIYNRSGND707 TYGAAHNLLV 717 AHALAWRLYD727 RQFRPSQRGA737 VSLSLHADWA747 EPANPYADSH757 WRAAERFLQF 767 EIAWFAEPLF777 KTGDYPAAMR787 EYIASKHRRG797 LSSSALPRLT807 EAERRLLKGT 817 VDFCALNHFT827 TRFVMHEQLA837 GSRYDSDRDI847 QFLQDITRLS857 SPTRLAVIPW 867 GVRKLLRWVR877 RNYGDMDIYI887 TASGIDDQAL897 EDDRLRKYYL907 GKYLQEVLKA 917 YLIDKVRIKG927 YYAFKLAEEK937 SKPRFGFFTS947 DFKAKSSIQF957 YNKVISSRGF 967 P
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Thermogenesis | hsa04714 | Affiliated Target |
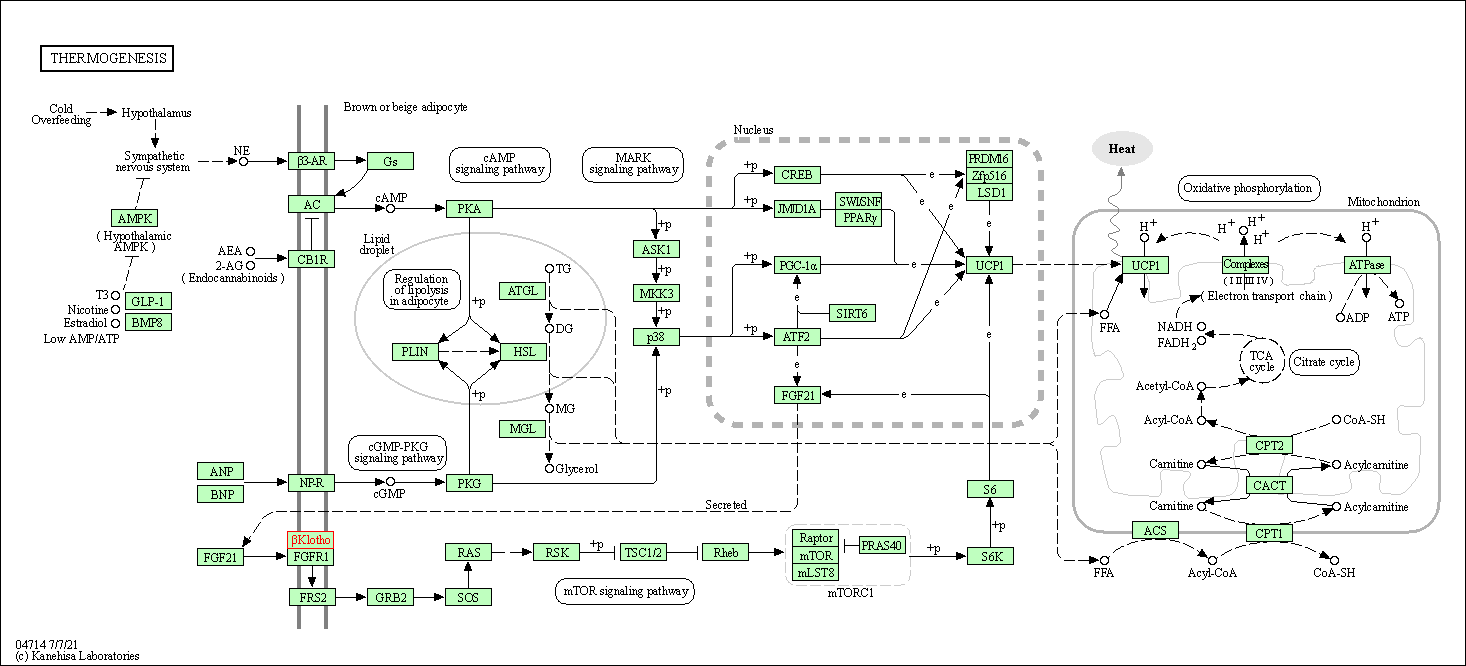
|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Degree | 7 | Degree centrality | 7.52E-04 | Betweenness centrality | 8.68E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.19E-01 | Radiality | 1.38E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 2.59E+01 | Topological coefficient | 1.98E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT04583423) A Phase 2b Randomized, Double-Blind, Placebo-Controlled Study to Evaluate the Efficacy and Safety of MK-3655 in Individuals With Pre-cirrhotic Nonalcoholic Steatohepatitis. U.S.National Institutes of Health. | |||||
| REF 3 | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling. Nature. 2018 Jan 25;553(7689):501-505. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

