Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T17980
(Former ID: TTDS00406)
|
|||||
| Target Name |
Fyn tyrosine protein kinase (FYN)
|
|||||
| Synonyms |
Tyrosine-protein kinase Fyn; Src-like kinase; SLK; Proto-oncogene tyrosine-protein kinase Fyn; Proto-oncogene c-Fyn; Proto-oncogene Syn; Fyn p59-Fyn; Fyn Protooncogene Syn
Click to Show/Hide
|
|||||
| Gene Name |
FYN
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Myeloproliferative neoplasm [ICD-11: 2A20] | |||||
| Function |
Inactive FYN is phosphorylated on its C-terminal tail within the catalytic domain. Following activation by PKA, the protein subsequently associates with PTK2/FAK1, allowing PTK2/FAK1 phosphorylation, activation and targeting to focal adhesions. Involved in the regulation of cell adhesion and motility through phosphorylation of CTNNB1 (beta-catenin) and CTNND1 (delta-catenin). Regulates cytoskeletal remodeling by phosphorylating several proteins including the actin regulator WAS and the microtubule-associated proteins MAP2 and MAPT. Promotes cell survival by phosphorylating AGAP2/PIKE-A and preventing its apoptotic cleavage. Participates in signal transduction pathways that regulate the integrity of the glomerular slit diaphragm (an essential part of the glomerular filter of the kidney) by phosphorylating several slit diaphragm components including NPHS1, KIRREL1 and TRPC6. Plays a role in neural processes by phosphorylating DPYSL2, a multifunctional adapter protein within the central nervous system, ARHGAP32, a regulator for Rho family GTPases implicated in various neural functions, and SNCA, a small pre-synaptic protein. Participates in the downstream signaling pathways that lead to T-cell differentiation and proliferation following T-cell receptor (TCR) stimulation. Phosphorylates PTK2B/PYK2 in response to T-cell receptor activation. Also participates in negative feedback regulation of TCR signaling through phosphorylation of PAG1, thereby promoting interaction between PAG1 and CSK and recruitment of CSK to lipid rafts. CSK maintains LCK and FYN in an inactive form. Promotes CD28-induced phosphorylation of VAV1. Non-receptor tyrosine-protein kinase that plays a role in many biological processes including regulation of cell growth and survival, cell adhesion, integrin-mediated signaling, cytoskeletal remodeling, cell motility, immune response and axon guidance.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.10.2
|
|||||
| Sequence |
MGCVQCKDKEATKLTEERDGSLNQSSGYRYGTDPTPQHYPSFGVTSIPNYNNFHAAGGQG
LTVFGGVNSSSHTGTLRTRGGTGVTLFVALYDYEARTEDDLSFHKGEKFQILNSSEGDWW EARSLTTGETGYIPSNYVAPVDSIQAEEWYFGKLGRKDAERQLLSFGNPRGTFLIRESET TKGAYSLSIRDWDDMKGDHVKHYKIRKLDNGGYYITTRAQFETLQQLVQHYSERAAGLCC RLVVPCHKGMPRLTDLSVKTKDVWEIPRESLQLIKRLGNGQFGEVWMGTWNGNTKVAIKT LKPGTMSPESFLEEAQIMKKLKHDKLVQLYAVVSEEPIYIVTEYMNKGSLLDFLKDGEGR ALKLPNLVDMAAQVAAGMAYIERMNYIHRDLRSANILVGNGLICKIADFGLARLIEDNEY TARQGAKFPIKWTAPEAALYGRFTIKSDVWSFGILLTELVTKGRVPYPGMNNREVLEQVE RGYRMPCPQDCPISLHELMIHCWKKDPEERPTFEYLQSFLEDYFTATEPQYQPGENL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T68CKY | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Phosphonotyrosine | Ligand Info | |||||
| Structure Description | Crystal Structure of Fyn kinase domain complexed with staurosporine | PDB:2DQ7 | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [11] |
| PDB Sequence |
KDVWEIPRES
10 LQLIKRLGNG20 QFGEVWMGTW30 NGNTKVAIKT40 LKPGTMSPES50 FLEEAQIMKK 60 LKHDKLVQLY70 AVVSEEPIYI80 VTEYMNKGSL90 LDFLKDGEGR100 ALKLPNLVDM 110 AAQVAAGMAY120 IERMNYIHRD130 LRSANILVGN140 GLICKIADFG150 LARLIEDNET 161 ARQGAKFPIK171 WTAPEAALYG181 RFTIKSDVWS191 FGILLTELVT201 KGRVPYPGMN 211 NREVLEQVER221 GYRMPCPQDC231 PISLHELMIH241 CWKKDPEERP251 TFEYLQSFLE 261 DY
|
|||||
|
|
||||||
| Ligand Name: Staurosporine | Ligand Info | |||||
| Structure Description | Crystal Structure of Fyn kinase domain complexed with staurosporine | PDB:2DQ7 | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [11] |
| PDB Sequence |
KDVWEIPRES
10 LQLIKRLGNG20 QFGEVWMGTW30 NGNTKVAIKT40 LKPGTMSPES50 FLEEAQIMKK 60 LKHDKLVQLY70 AVVSEEPIYI80 VTEYMNKGSL90 LDFLKDGEGR100 ALKLPNLVDM 110 AAQVAAGMAY120 IERMNYIHRD130 LRSANILVGN140 GLICKIADFG150 LARLIEDNET 161 ARQGAKFPIK171 WTAPEAALYG181 RFTIKSDVWS191 FGILLTELVT201 KGRVPYPGMN 211 NREVLEQVER221 GYRMPCPQDC231 PISLHELMIH241 CWKKDPEERP251 TFEYLQSFLE 261 DY
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |
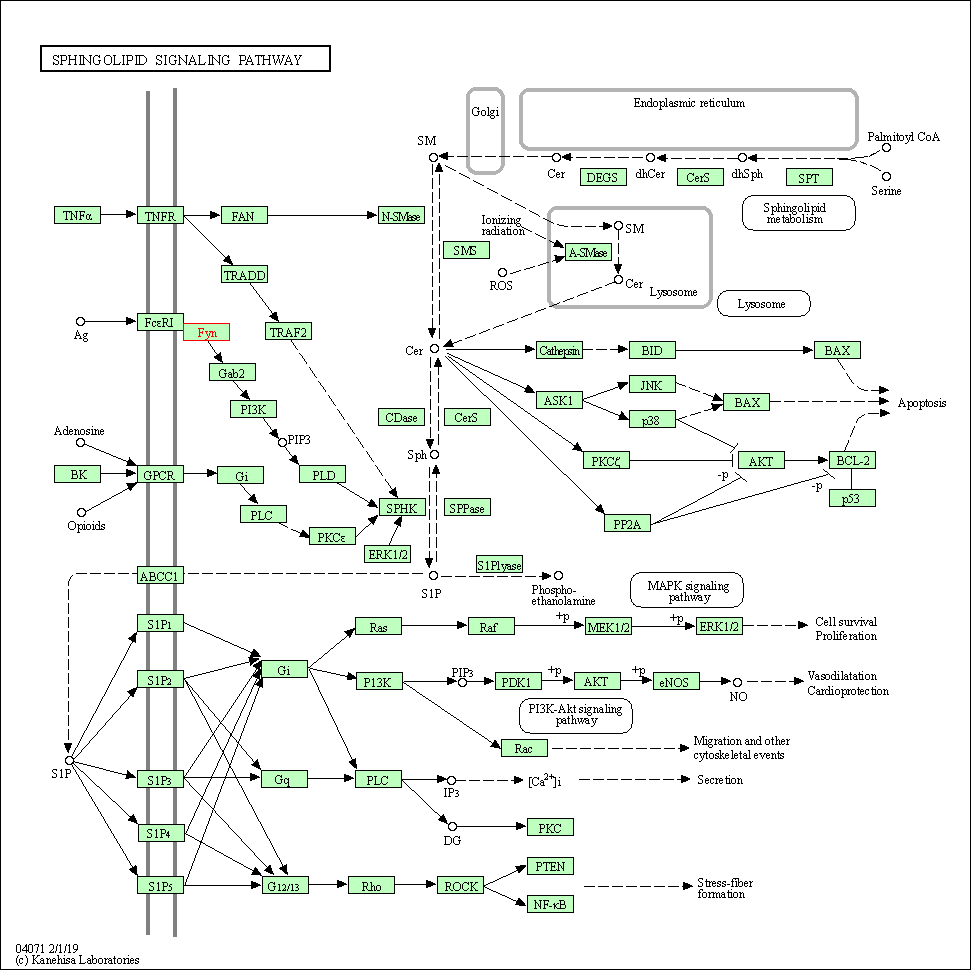
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |
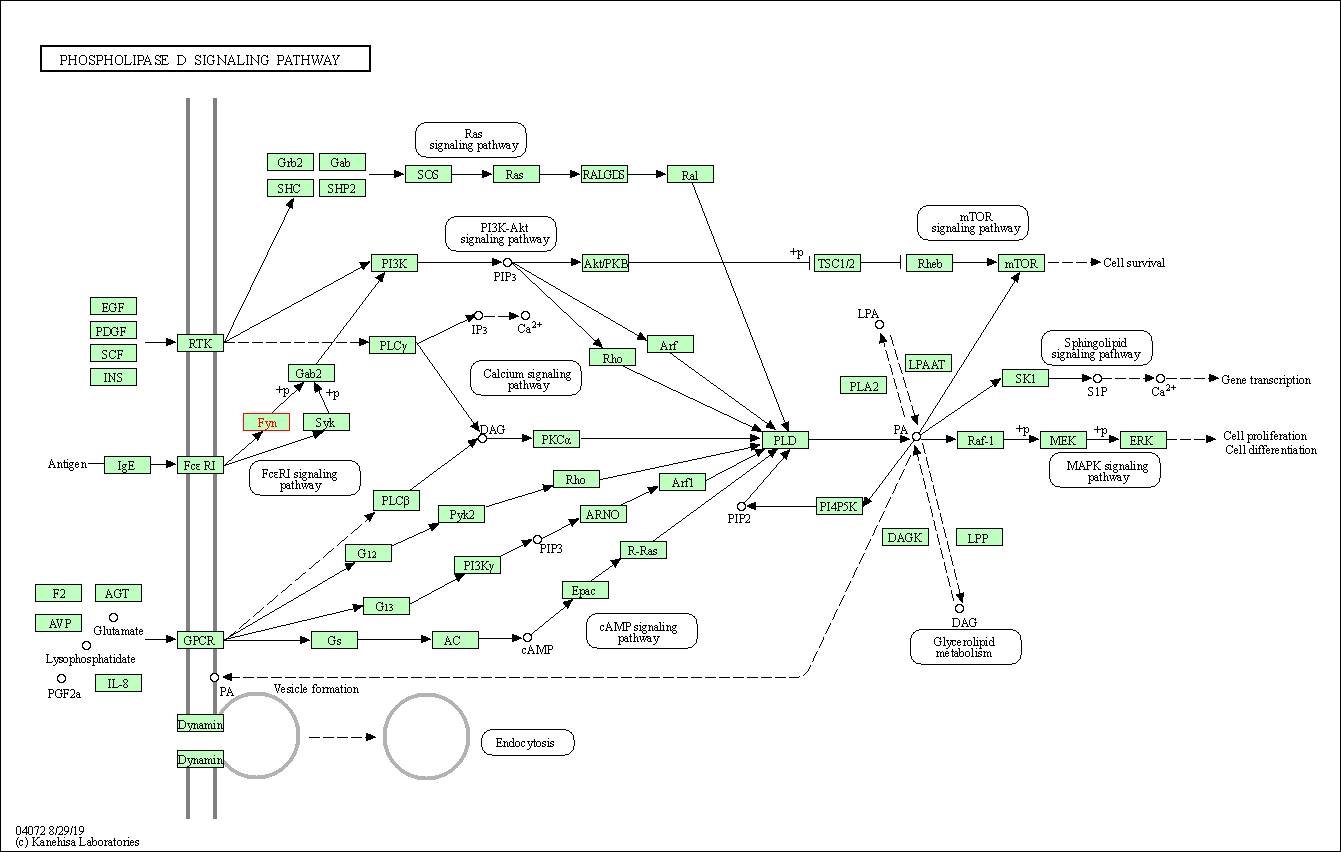
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Axon guidance | hsa04360 | Affiliated Target |
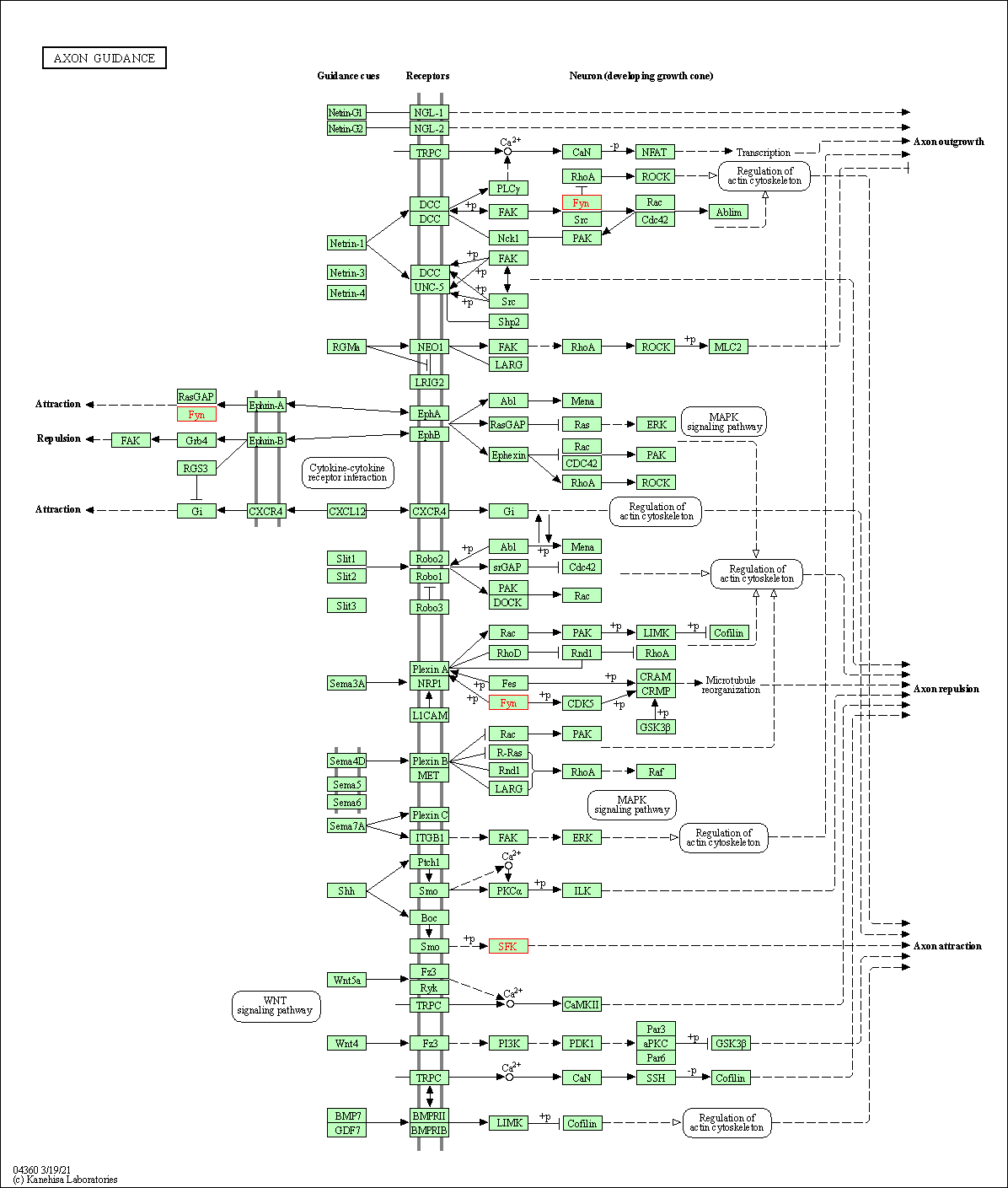
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |
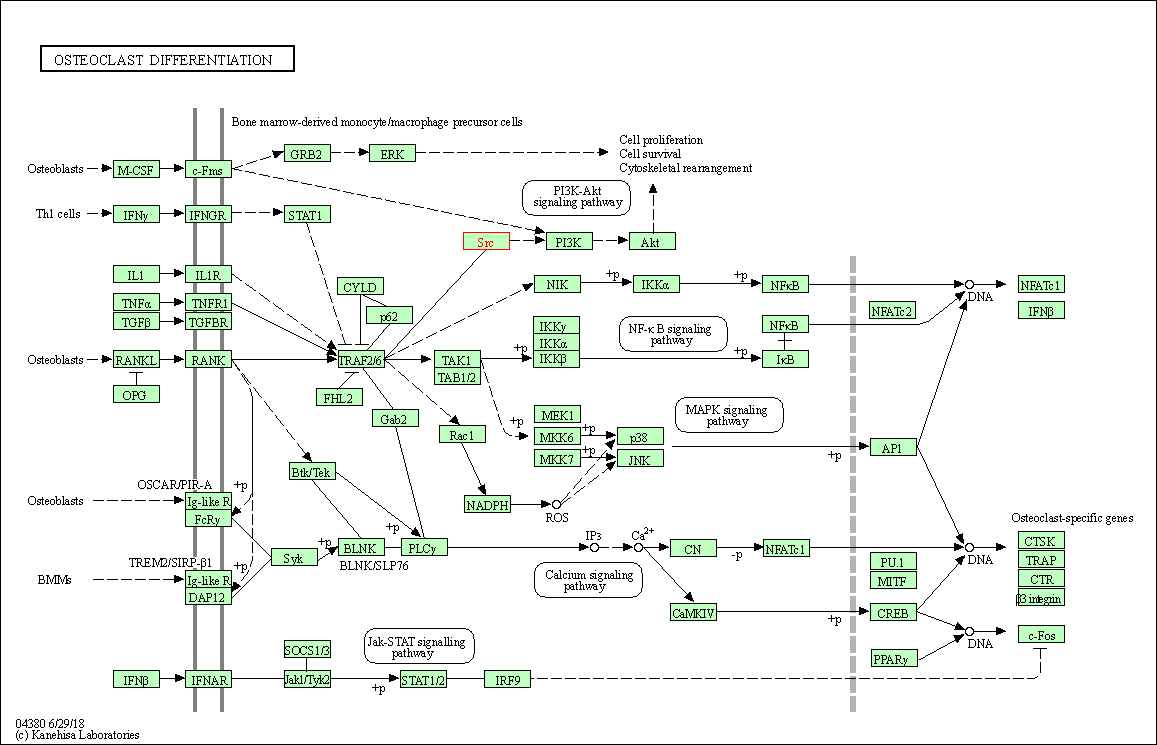
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |
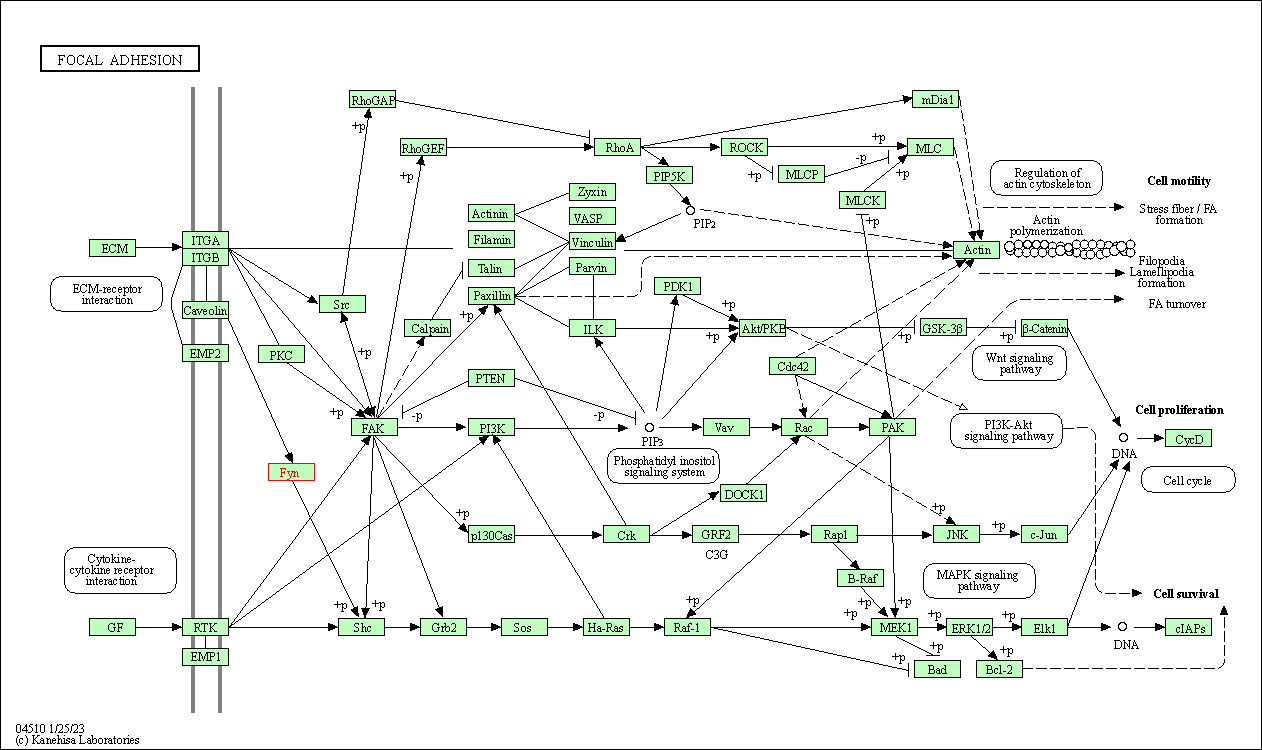
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Adherens junction | hsa04520 | Affiliated Target |
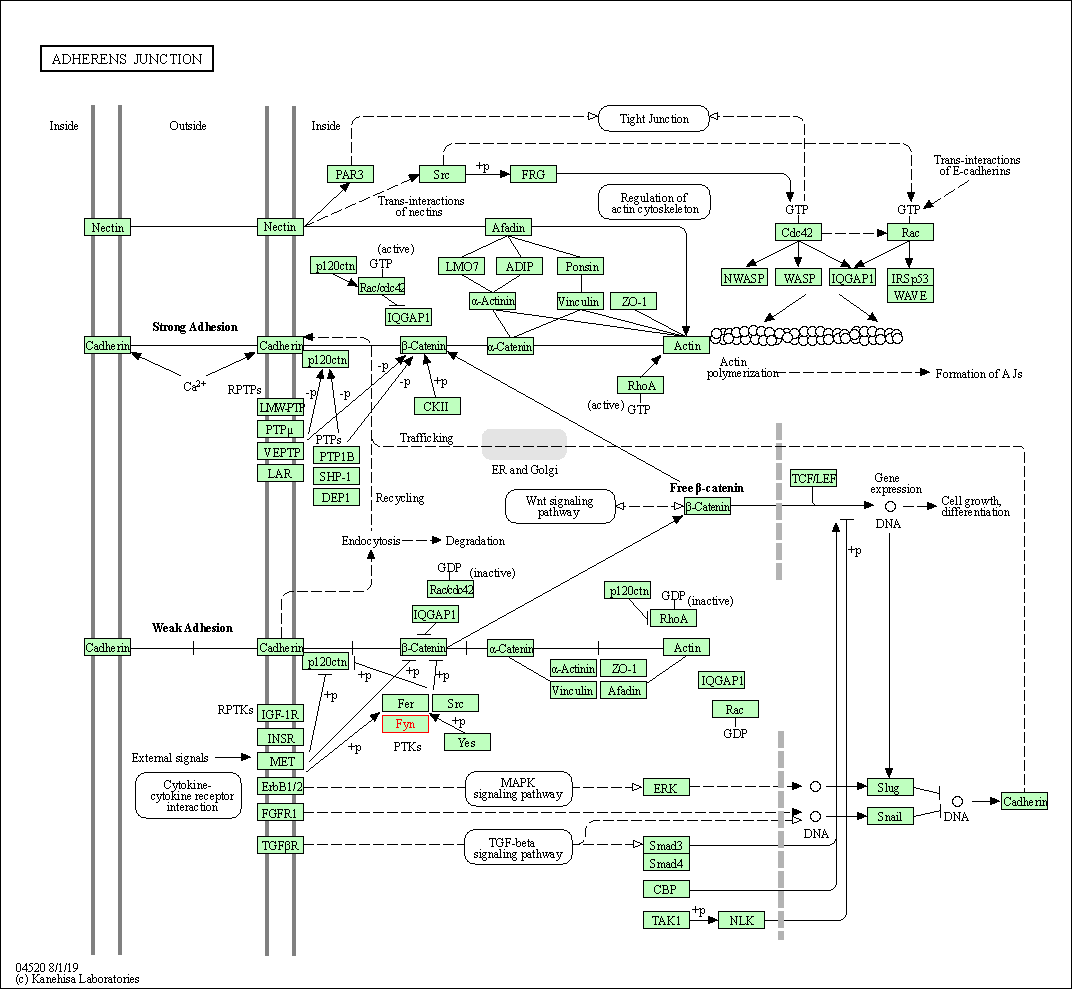
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |
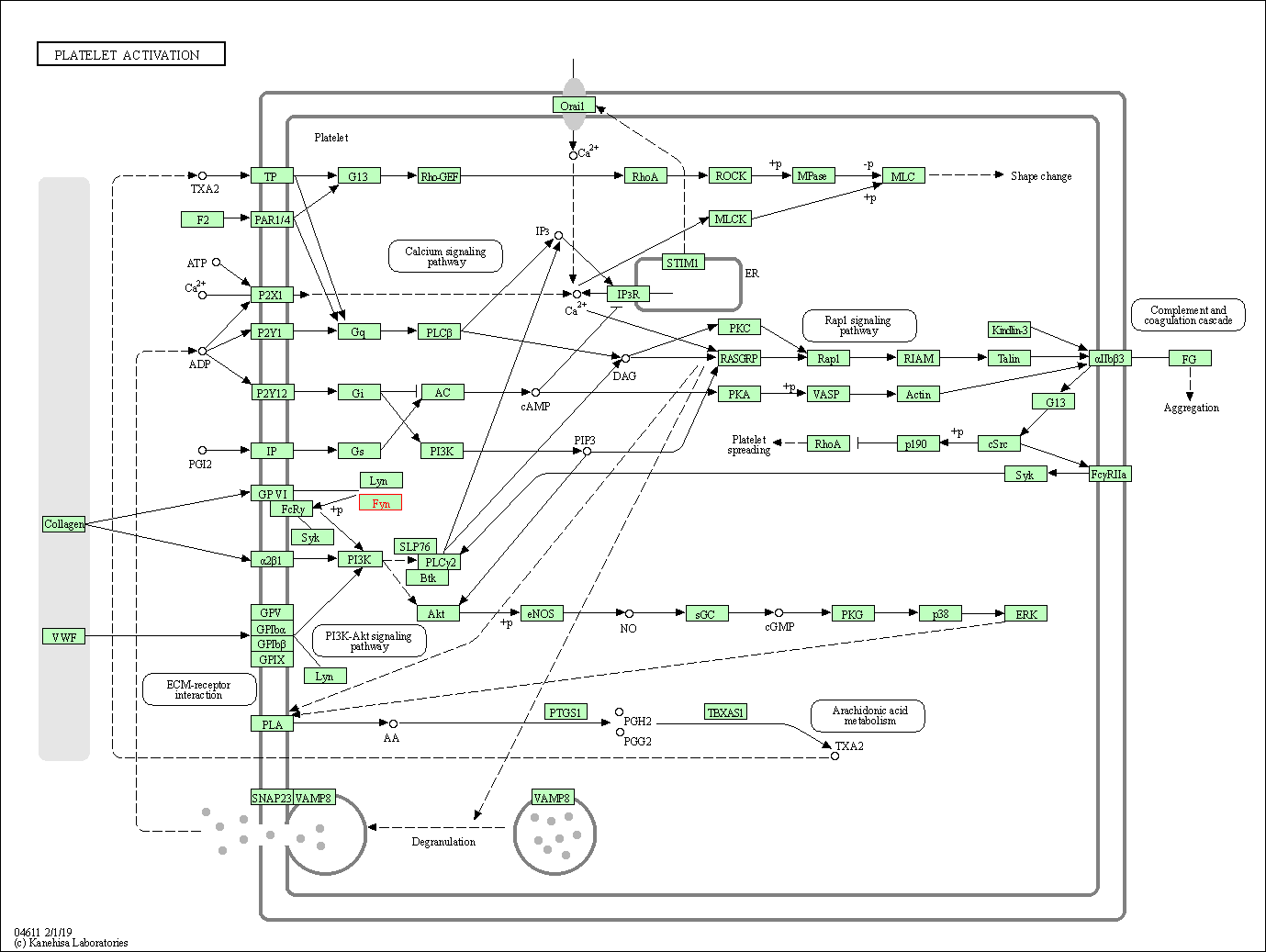
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |
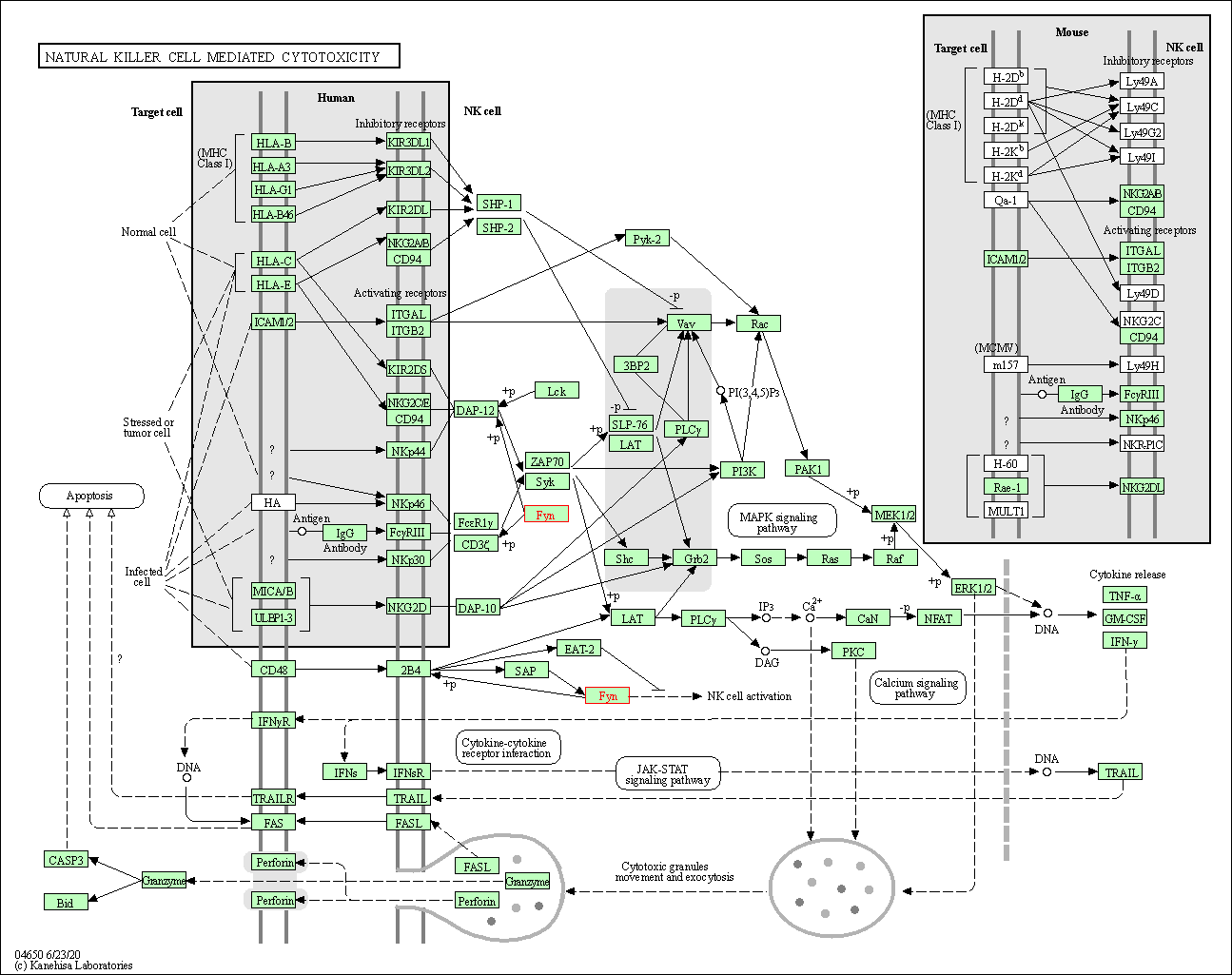
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |
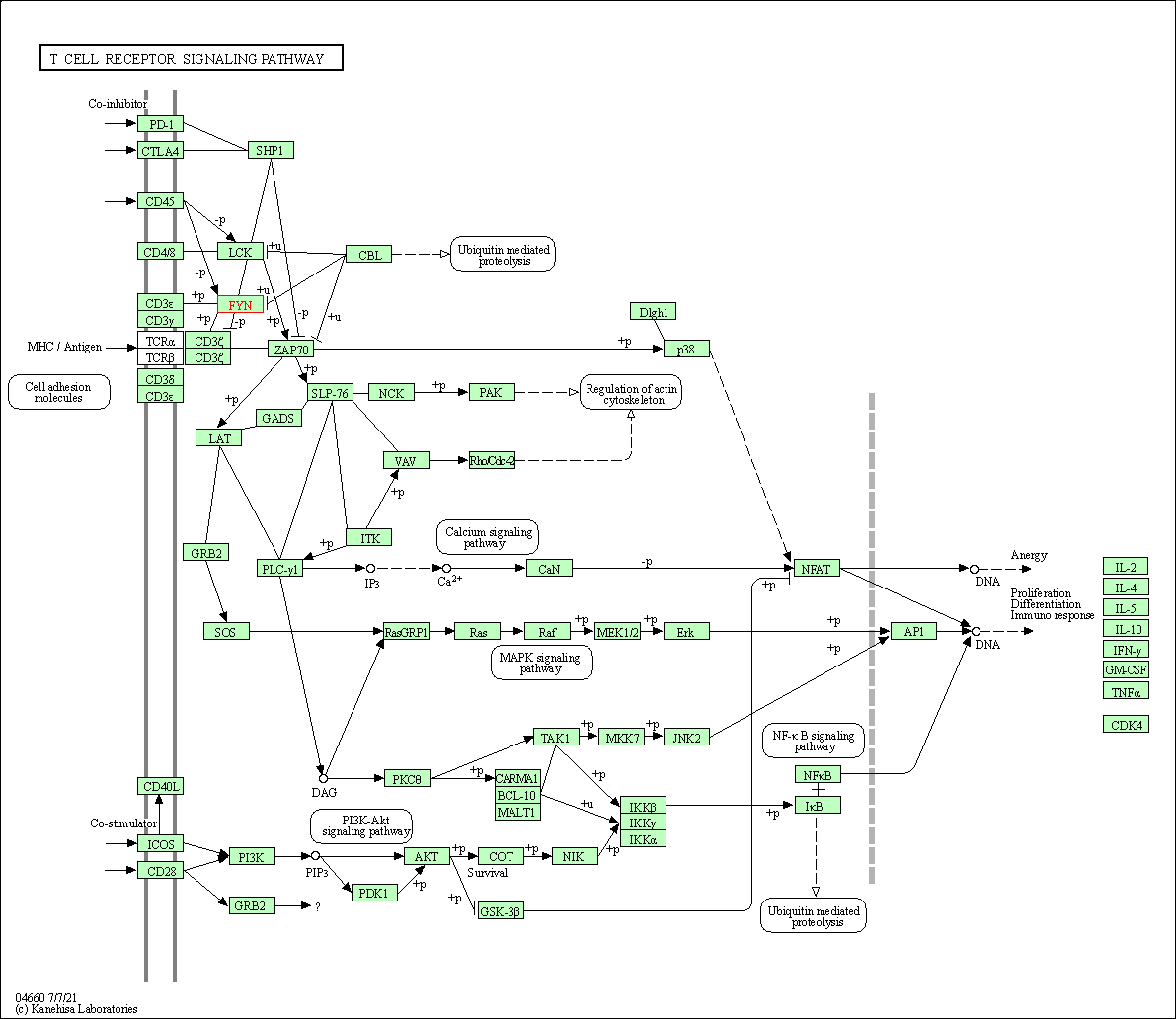
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |
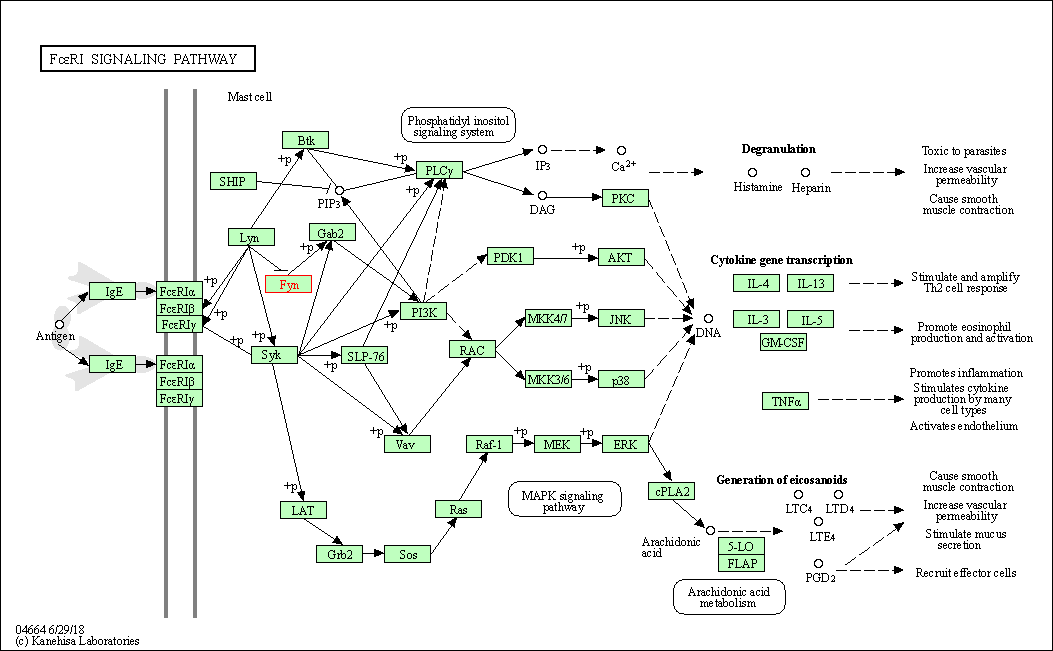
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Cholinergic synapse | hsa04725 | Affiliated Target |
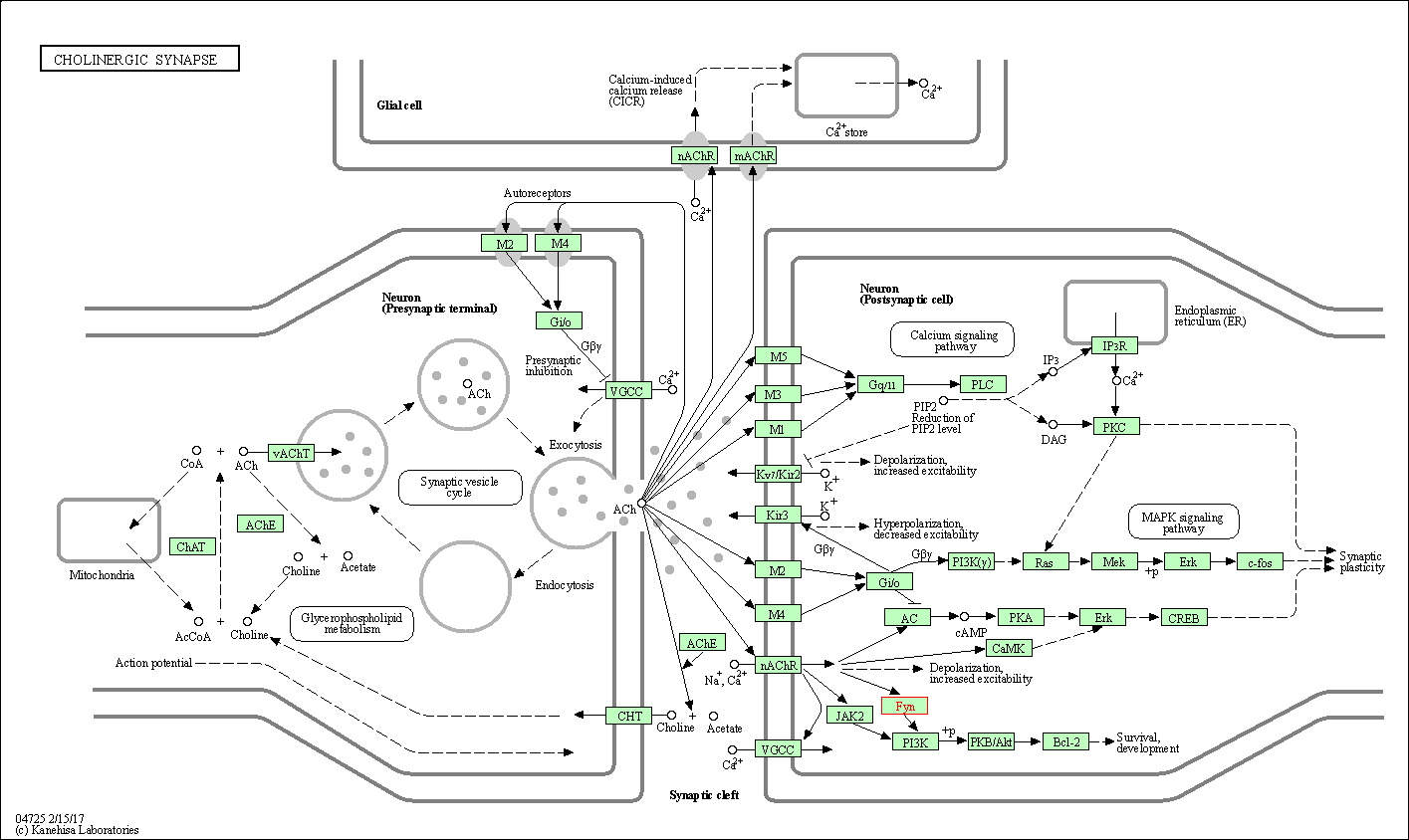
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 87 | Degree centrality | 9.35E-03 | Betweenness centrality | 1.00E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.71E-01 | Radiality | 1.46E+01 | Clustering coefficient | 1.09E-01 |
| Neighborhood connectivity | 4.10E+01 | Topological coefficient | 2.97E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | A comparison of physicochemical property profiles of marketed oral drugs and orally bioavailable anti-cancer protein kinase inhibitors in clinical development. Curr Top Med Chem. 2007;7(14):1408-22. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5678). | |||||
| REF 3 | 2006 drug approvals: finding the niche. Nat Rev Drug Discov. 2007 Feb;6(2):99-101. | |||||
| REF 4 | Dasatinib: a tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia and philadelphia chromosome-positive acute lymphoblastic leukemia. Clin Ther. 2007 Nov;29(11):2289-308. | |||||
| REF 5 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 6 | ClinicalTrials.gov (NCT00676299) A Safety and Dose-finding Study of JNJ-26483327, a Drug in Development for Cancer, for Patients With Advanced and/or Refractory Solid Malignancies.. U.S. National Institutes of Health. | |||||
| REF 7 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 8 | Mixed-lineage kinase 1 and mixed-lineage kinase 3 subtype-selective dihydronaphthyl[3,4-a]pyrrolo[3,4-c]carbazole-5-ones: optimization, mixed-linea... J Med Chem. 2008 Sep 25;51(18):5680-9. | |||||
| REF 9 | Discovery of novel 2-(aminoheteroaryl)-thiazole-5-carboxamides as potent and orally active Src-family kinase p56(Lck) inhibitors. Bioorg Med Chem Lett. 2004 Dec 20;14(24):6061-6. | |||||
| REF 10 | Biological evaluation of a multi-targeted small molecule inhibitor of tumor-induced angiogenesis. Bioorg Med Chem Lett. 2006 Apr 1;16(7):1950-3. | |||||
| REF 11 | Structure of human Fyn kinase domain complexed with staurosporine. Biochem Biophys Res Commun. 2006 Aug 4;346(3):840-4. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

