Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T19229
(Former ID: TTDI03501)
|
|||||
| Target Name |
Proton-coupled folate transporter (SLC46A1)
|
|||||
| Synonyms |
Solute carrier family 46 member 1; PCFT/HCP1; PCFT; Heme carrier protein 1; HCP1; G21
Click to Show/Hide
|
|||||
| Gene Name |
SLC46A1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Folate deficiency anaemia [ICD-11: 3A02] | |||||
| 2 | Leukaemia [ICD-11: 2A60-2B33] | |||||
| 3 | Nutritional deficiency [ICD-11: 5B50-5B71] | |||||
| Function |
Has been shown to act both as an intestinal proton-coupled high-affinity folate transporter and as an intestinal heme transporter which mediates heme uptake from the gut lumen into duodenal epithelial cells. The iron is then released from heme and may be transported into the bloodstream. Dietary heme iron is an important nutritional source of iron. Shows a higher affinity for folate than heme.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MEGSASPPEKPRARPAAAVLCRGPVEPLVFLANFALVLQGPLTTQYLWHRFSADLGYNGT
RQRGGCSNRSADPTMQEVETLTSHWTLYMNVGGFLVGLFSSTLLGAWSDSVGRRPLLVLA SLGLLLQALVSVFVVQLQLHVGYFVLGRILCALLGDFGGLLAASFASVADVSSSRSRTFR MALLEASIGVAGMLASLLGGHWLRAQGYANPFWLALALLIAMTLYAAFCFGETLKEPKST RLFTFRHHRSIVQLYVAPAPEKSRKHLALYSLAIFVVITVHFGAQDILTLYELSTPLCWD SKLIGYGSAAQHLPYLTSLLALKLLQYCLADAWVAEIGLAFNILGMVVFAFATITPLMFT GYGLLFLSLVITPVIRAKLSKLVRETEQGALFSAVACVNSLAMLTASGIFNSLYPATLNF MKGFPFLLGAGLLLIPAVLIGMLEKADPHLEFQQFPQSP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Methotrexate | Drug Info | Approved | leukaemia | [5] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Modulator | [+] 3 Modulator drugs | + | ||||
| 1 | Methotrexate | Drug Info | [1] | |||
| 2 | [3H]folinic acid | Drug Info | [1] | |||
| 3 | [3H]N5-methylfolate | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Vitamin digestion and absorption | hsa04977 | Affiliated Target |
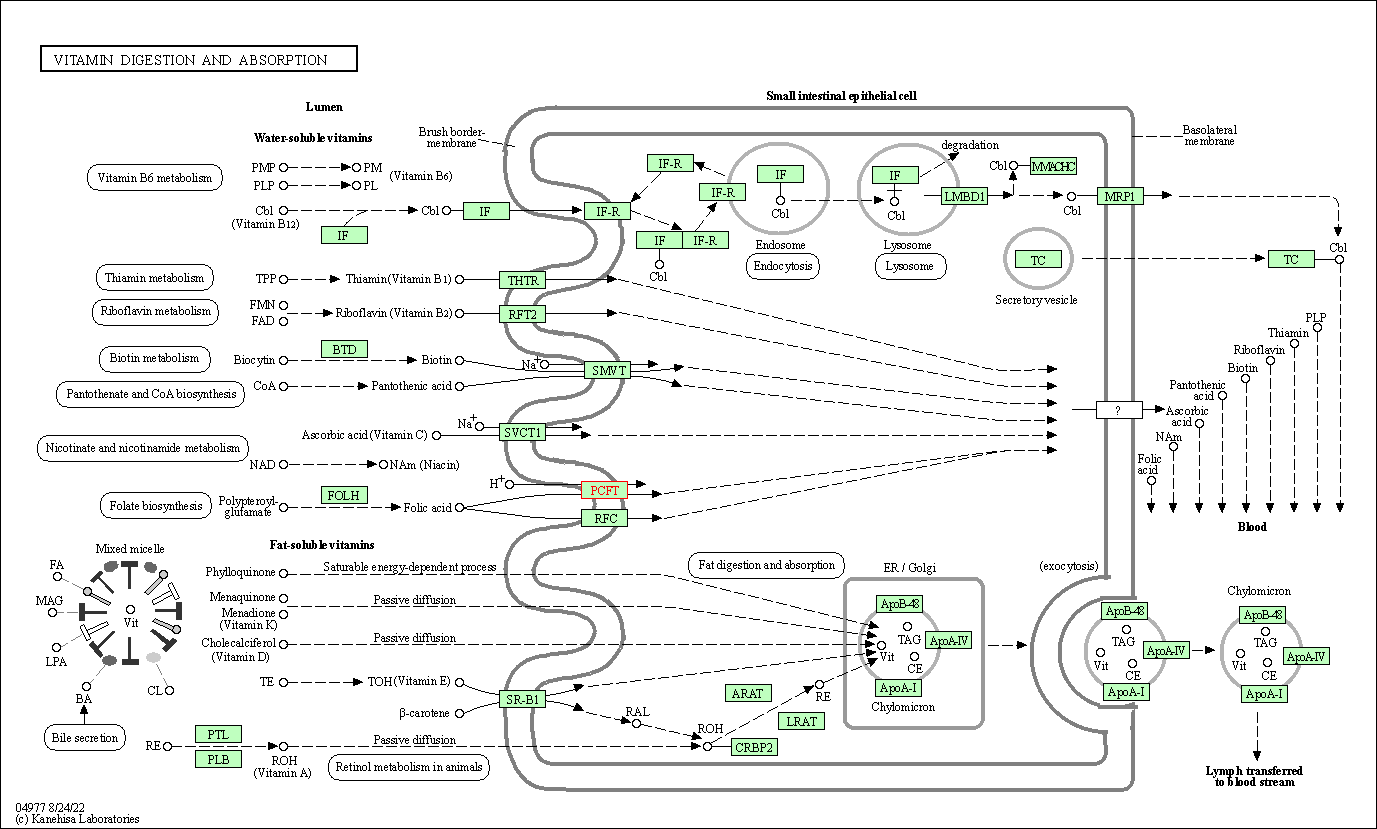
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Mineral absorption | hsa04978 | Affiliated Target |
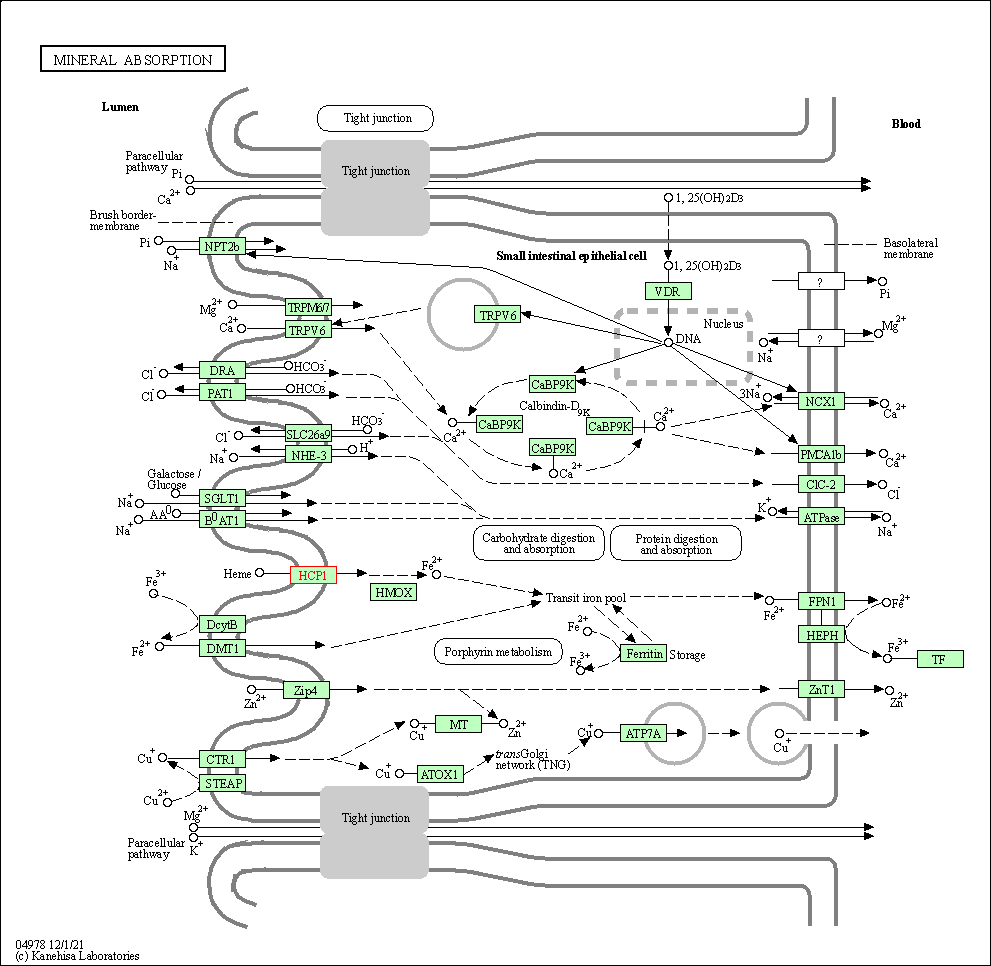
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1213). | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4563). | |||||
| REF 3 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (ANDA) 040514. | |||||
| REF 4 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 5 | Pain reduction with oral methotrexate in knee osteoarthritis, a pragmatic phase iii trial of treatment effectiveness (PROMOTE): study protocol for ... Trials. 2015 Mar 4;16:77. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

