Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T21307
(Former ID: TTDI02119)
|
|||||
| Target Name |
MAPK-activated protein kinase 2 (MAPKAPK2)
|
|||||
| Synonyms |
MK2; MK-2; MAPKactivated protein kinase 2; MAPKAPK-2; MAPKAP-K2; MAPKAP kinase 2; MAP kinaseactivated protein kinase 2; MAP kinase-activated protein kinase 2
Click to Show/Hide
|
|||||
| Gene Name |
MAPKAPK2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 7 Target-related Diseases | + | ||||
| 1 | Hidradenitis suppurativa [ICD-11: ED92] | |||||
| 2 | Monogenic autoinflammatory syndrome ICD-11: 4A60 | |||||
| 3 | Psoriatic arthritis [ICD-11: FA21] | |||||
| 4 | Rheumatoid arthritis [ICD-11: FA20] | |||||
| 5 | Peritoneal cancer [ICD-11: 2C51] | |||||
| 6 | Acute laryngitis/tracheitis [ICD-11: CA05] | |||||
| 7 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Following stress, it is phosphorylated and activated by MAP kinase p38-alpha/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. Phosphorylates ALOX5, CDC25B, CDC25C, CEP131, ELAVL1, HNRNPA0, HSP27/HSPB1, KRT18, KRT20, LIMK1, LSP1, PABPC1, PARN, PDE4A, RCSD1, RPS6KA3, TAB3 and TTP/ZFP36. Phosphorylates HSF1; leading to the interaction with HSP90 proteins and inhibiting HSF1 homotrimerization, DNA-binding and transactivation activities. Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to the dissociation of HSP27/HSPB1 from large small heat-shock protein (sHsps) oligomers and impairment of their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating tumor necrosis factor (TNF) and IL6 production post-transcriptionally: acts by phosphorylating AU-rich elements (AREs)-binding proteins ELAVL1, HNRNPA0, PABPC1 and TTP/ZFP36, leading to the regulation of the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major post-transcriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity, leading to inhibition of dependent degradation of ARE-containing transcripts. Phosphorylates CEP131 in response to cellular stress induced by ultraviolet irradiation which promotes binding of CEP131 to 14-3-3 proteins and inhibits formation of novel centriolar satellites. Also involved in late G2/M checkpoint following DNA damage through a process of post-transcriptional mRNA stabilization: following DNA damage, relocalizes from nucleus to cytoplasm and phosphorylates HNRNPA0 and PARN, leading to stabilization of GADD45A mRNA. Involved in toll-like receptor signaling pathway (TLR) in dendritic cells: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. Stress-activated serine/threonine-protein kinase involved in cytokine production, endocytosis, reorganization of the cytoskeleton, cell migration, cell cycle control, chromatin remodeling, DNA damage response and transcriptional regulation.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MLSNSQGQSPPVPFPAPAPPPQPPTPALPHPPAQPPPPPPQQFPQFHVKSGLQIKKNAII
DDYKVTSQVLGLGINGKVLQIFNKRTQEKFALKMLQDCPKARREVELHWRASQCPHIVRI VDVYENLYAGRKCLLIVMECLDGGELFSRIQDRGDQAFTEREASEIMKSIGEAIQYLHSI NIAHRDVKPENLLYTSKRPNAILKLTDFGFAKETTSHNSLTTPCYTPYYVAPEVLGPEKY DKSCDMWSLGVIMYILLCGYPPFYSNHGLAISPGMKTRIRMGQYEFPNPEWSEVSEEVKM LIRNLLKTEPTQRMTITEFMNHPWIMQSTKVPQTPLHTSRVLKEDKERWEDVKEEMTSAL ATMRVDYEQIKIKKIEDASNPLLLKRRKKARALEAAALAH Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T11B42 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | CBP-501 | Drug Info | Phase 1/2 | Mesothelioma | [3] | |
| 2 | MMI-0100 | Drug Info | Phase 1 | Airway inflammation | [4] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 6 Inhibitor drugs | + | ||||
| 1 | CBP-501 | Drug Info | [1] | |||
| 2 | MMI-0100 | Drug Info | [5] | |||
| 3 | CMPD1 | Drug Info | [6] | |||
| 4 | PF-3644022 | Drug Info | [7] | |||
| 5 | PMID17480064C16 | Drug Info | [8] | |||
| 6 | PMID19364658C33 | Drug Info | [9] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Staurosporine | Ligand Info | |||||
| Structure Description | Crystal structure of staurosporine bound to MAP KAP kinase 2 | PDB:1NXK | ||||
| Method | X-ray diffraction | Resolution | 2.70 Å | Mutation | Yes | [10] |
| PDB Sequence |
QFPQFHVKSG
51 LQIKKNAIID61 DYKVTSQVLG71 LGINGKVLQI81 FNKRTQEKFA91 LKLQDCPKAR 102 REVELHWRAS112 QCPHIVRIVD122 VYENLYAGRK132 CLLIVECLDG143 GELFSRIQDR 153 AFTEREASEI166 KSIGEAIQYL177 HSINIAHRDV187 KPENLLYTSK197 RPNAILKLTD 207 FGFAKETTPY228 YVAPEVLGPE238 KYDKSCDWSL249 GVIYILLCGY260 PPFYSNHGLA 270 ISPGKTRIRG282 QYEFPNPEWS292 EVSEEVKLIR303 NLLKTEPTQR313 TITEFNHPWI 325 QSTKVPQTPL336 HTSRVLKED
|
|||||
|
|
||||||
| Ligand Name: adenosine diphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of ADP bound to MAP KAP kinase 2 | PDB:1NY3 | ||||
| Method | X-ray diffraction | Resolution | 3.00 Å | Mutation | No | [10] |
| PDB Sequence |
FHVKSGLQIK
55 KNAIIDDYKV65 TSQVLGLGIN75 GKVLQIFNKR85 TQEKFALKML95 QDCPKARREV 105 ELHWRASQCP115 HIVRIVDVYE125 NLYAGRKCLL135 IVMECLDGGE145 LFSRIQDTER 161 EASEIMKSIG171 EAIQYLHSIN181 IAHRDVKPEN191 LLYTSKRPNA201 ILKLTDFGFA 211 KETTSHPYYV230 APEVLGPEKY240 DKSCDMWSLG250 VIMYILLCGY260 PPFYSGMKTR 278 IRMGQYEFPN288 PEWSEVSEEV298 KMLIRNLLKT308 EPTQRMTITE318 FMNHPWIMQS 328 TKVPQTPLHT338 SRVLKED
|
|||||
|
|
LEU70
3.663
GLY71
3.643
LEU72
3.542
GLY73
2.809
ILE74
3.052
ASN75
3.763
GLY76
4.874
VAL78
3.710
ALA91
3.456
LYS93
2.641
VAL118
3.967
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |
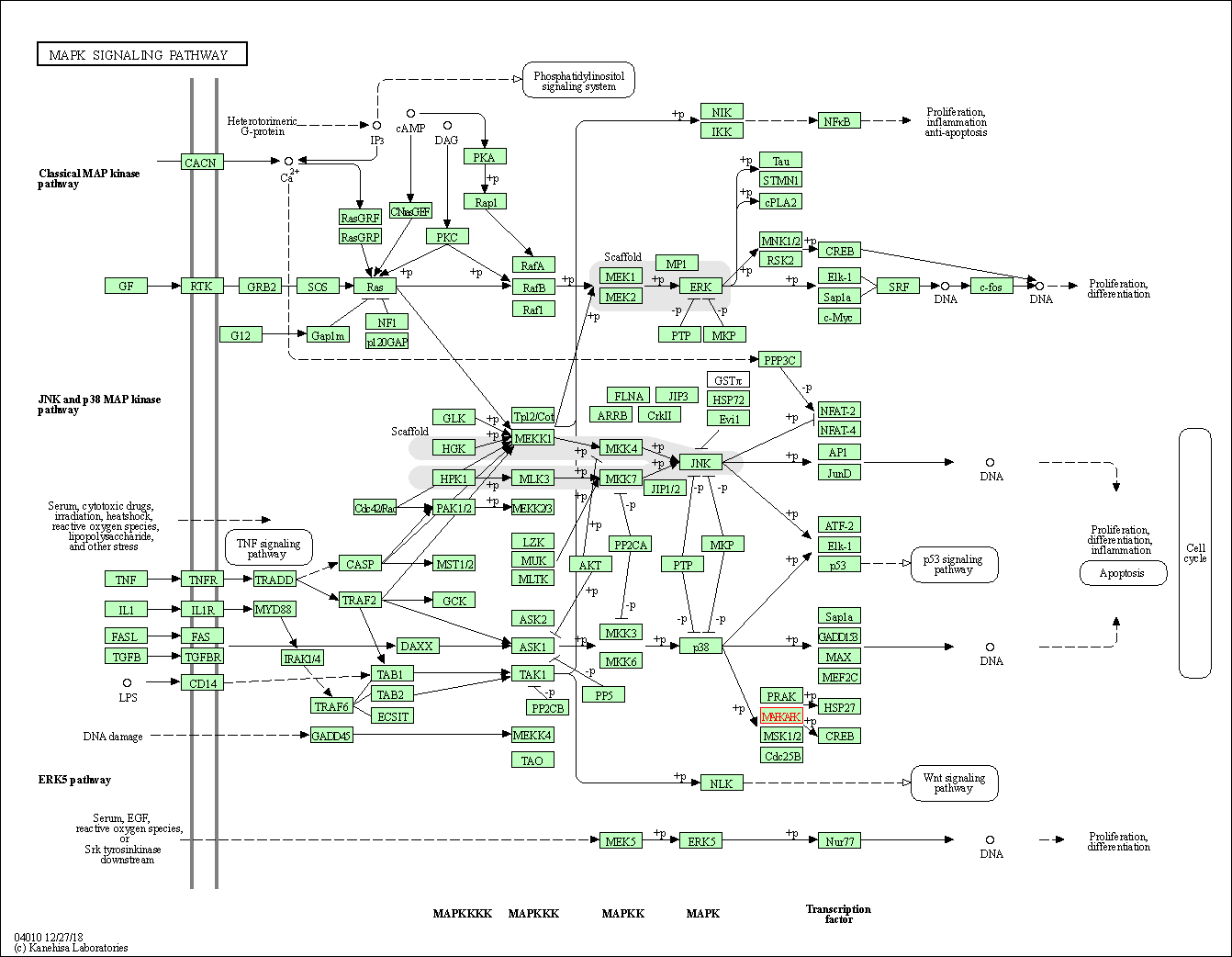
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
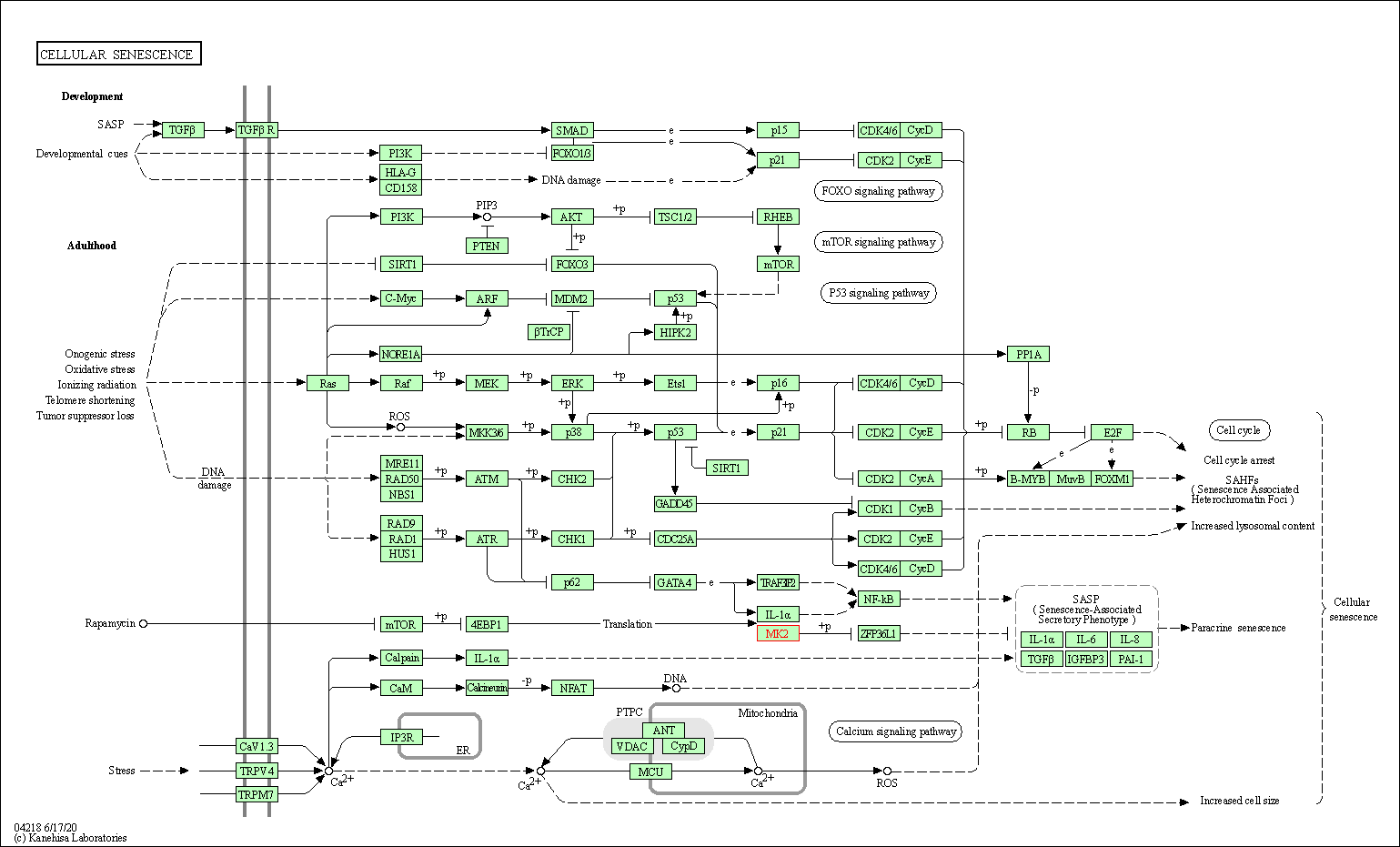
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| VEGF signaling pathway | hsa04370 | Affiliated Target |
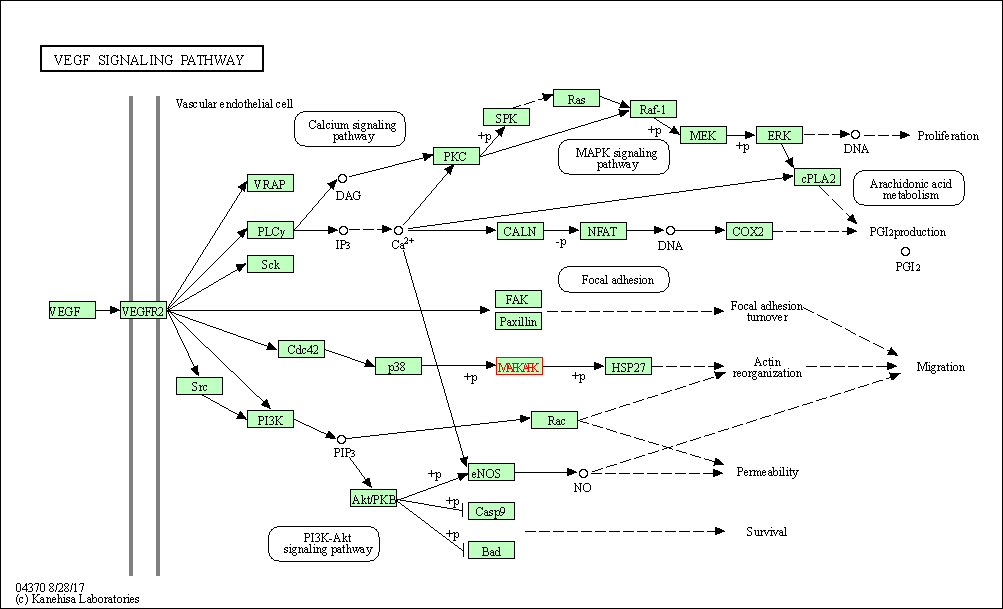
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 5.16E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.17E-01 | Radiality | 1.38E+01 | Clustering coefficient | 1.07E-01 |
| Neighborhood connectivity | 1.94E+01 | Topological coefficient | 1.64E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | National Cancer Institute Drug Dictionary (drug id 577812). | |||||
| REF 2 | ClinicalTrials.gov (NCT05511519) A Phase 2a, Randomized, Double-blind, Placebo-controlled Study to Investigate the Efficacy, Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of Zunsemetinib vs Placebo in Patients With Moderate-to-Severe Active Psoriatic Arthritis. U.S.National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT00700336) Study of CBP501 + Pemetrexed + Cisplatin on MPM (Phase I/II). U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT02515396) Phase 1a Study in Healthy Smokers to Investigate the Effect With MMI-0100 on Airway Inflammation in Induced Sputum After Challenge With Inhaled Lipopolysaccharide (LPS). U.S. National Institutes of Health. | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2094). | |||||
| REF 6 | Synthesis and in vivo activity of MK2 and MK2 substrate-selective p38alpha(MAPK) inhibitors in Werner syndrome cells. Bioorg Med Chem Lett. 2007 Dec 15;17(24):6832-5. | |||||
| REF 7 | A benzothiophene inhibitor of mitogen-activated protein kinase-activated protein kinase 2 inhibits tumor necrosis factor alpha production and has oral anti-inflammatory efficacy in acute and chronic models of inflammation. J Pharmacol Exp Ther. 2010 Jun;333(3):797-807. | |||||
| REF 8 | Pyrrolopyridine inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2). J Med Chem. 2007 May 31;50(11):2647-54. | |||||
| REF 9 | Identification and SAR of squarate inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem. 2009 May 1;17(9):3342-51. | |||||
| REF 10 | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme. Structure. 2003 Jun;11(6):627-36. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

