Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T23471
(Former ID: TTDC00199)
|
|||||
| Target Name |
Myeloperoxidase (MPO)
|
|||||
| Synonyms |
MPO
Click to Show/Hide
|
|||||
| Gene Name |
MPO
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Postoperative inflammation [ICD-11: 1A00-CA43] | |||||
| 2 | Left ventricular failure ICD-11: BD11 | |||||
| Function |
Part of the host defense system of polymorphonuclear leukocytes. It is responsible for microbicidal activity against a wide range of organisms. In the stimulated PMN, MPO catalyzes the production of hypohalous acids, primarily hypochlorous acidin physiologic situations, and other toxic intermediates that greatly enhance PMN microbicidal activity.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peroxidases
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.11.2.2
|
|||||
| Sequence |
MGVPFFSSLRCMVDLGPCWAGGLTAEMKLLLALAGLLAILATPQPSEGAAPAVLGEVDTS
LVLSSMEEAKQLVDKAYKERRESIKQRLRSGSASPMELLSYFKQPVAATRTAVRAADYLH VALDLLERKLRSLWRRPFNVTDVLTPAQLNVLSKSSGCAYQDVGVTCPEQDKYRTITGMC NNRRSPTLGASNRAFVRWLPAEYEDGFSLPYGWTPGVKRNGFPVALARAVSNEIVRFPTD QLTPDQERSLMFMQWGQLLDHDLDFTPEPAARASFVTGVNCETSCVQQPPCFPLKIPPND PRIKNQADCIPFFRSCPACPGSNITIRNQINALTSFVDASMVYGSEEPLARNLRNMSNQL GLLAVNQRFQDNGRALLPFDNLHDDPCLLTNRSARIPCFLAGDTRSSEMPELTSMHTLLL REHNRLATELKSLNPRWDGERLYQEARKIVGAMVQIITYRDYLPLVLGPTAMRKYLPTYR SYNDSVDPRIANVFTNAFRYGHTLIQPFMFRLDNRYQPMEPNPRVPLSRVFFASWRVVLE GGIDPILRGLMATPAKLNRQNQIAVDEIRERLFEQVMRIGLDLPALNMQRSRDHGLPGYN AWRRFCGLPQPETVGQLGTVLRNLKLARKLMEQYGTPNNIDIWMGGVSEPLKRKGRVGPL LACIIGTQFRKLRDGDRFWWENEGVFSMQQRQALAQISLPRIICDNTGITTVSKNNIFMS NSYPRDFVNCSTLPALNLASWREAS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A00127 ; BADD_A01943 ; BADD_A02053 ; BADD_A02603 ; BADD_A03271 ; BADD_A03824 ; BADD_A04537 ; BADD_A04561 ; BADD_A06206 ; BADD_A07267 | |||||
| HIT2.0 ID | T30VNV | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | AZD-3241 | Drug Info | Phase 2 | Parkinson disease | [4] | |
| 2 | AZD4831 | Drug Info | Phase 1 | Heart failure | [5] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 3 Inhibitor drugs | + | ||||
| 1 | AZD-3241 | Drug Info | [8] | |||
| 2 | 4-aminobenzoic acid hydrazide | Drug Info | [9] | |||
| 3 | INV-311 | Drug Info | [10] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | AZD4831 | Drug Info | [5] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Paroxetine | Ligand Info | |||||
| Structure Description | Glycosylation in the crystal structure of neutrophil myeloperoxidase | PDB:7OIH | ||||
| Method | X-ray diffraction | Resolution | 2.60 Å | Mutation | No | [11] |
| PDB Sequence |
TCPEQDKYRT
175 ITGMCNNRRS185 PTLGASNRAF195 VRWLPAEYED205 GFSLPYGWTP215 GVKRNGFPVA 225 LARAVSNEIV235 RFPTDQLTPD245 QERSLMFMQW255 GQLLDHDLDF265 TPEPAARVNC 281 ETSCVQQPPC291 FPLKIPPNDP301 RIKNQADCIP311 FFRSPACPGS322 NITIRNQINA 332 LTSFVDASMV342 YGSEEPLARN352 LRNMSNQLGL362 LAVNQRFQDN372 GRALLPFDNL 382 HDDPCLLTNR392 SARIPCFLAG402 DTRSSEMPEL412 TSMHTLLLRE422 HNRLATELKS 432 LNPRWDGERL442 YQEARKIVGA452 MVQIITYRDY462 LPLVLGPTAM472 RKYLPTYRSY 482 NDSVDPRIAN492 VFTNAFRYGH502 TLIQPFMFRL512 DNRYQPMEPN522 PRVPLSRVFF 532 ASWRVVLEGG542 IDPILRGLMA552 TPAKLNRQNQ562 IAVDEIRERL572 FEQVMRIGLD 582 LPALNMQRSR592 DHGLPGYNAW602 RRFCGLPQPE612 TVGQLGTVLR622 NLKLARKLME 632 QYGTPNNIDI642 WMGGVSEPLK652 RKGRVGPLLA662 CIIGTQFRKL672 RDGDRFWWEN 682 EGVFSMQQRQ692 ALAQISLPRI702 ICDNTGITTV712 SKNNIFMSNS722 YPRDFVNCST 732 LPALNLASWR742 EA
|
|||||
|
|
||||||
| Ligand Name: Beta-D-Mannose | Ligand Info | |||||
| Structure Description | A Structurally Dynamic N-terminal Region Drives Function of the Staphylococcal Peroxidase Inhibitor (SPIN) | PDB:6AZP | ||||
| Method | X-ray diffraction | Resolution | 2.29 Å | Mutation | No | [12] |
| PDB Sequence |
CPEQDKYRTI
176 TGMCNNRRSP186 TLGASNRAFV196 RWLPAEYEDG206 FSLPYGWTPG216 VKRNGFPVAL 226 ARAVSNEIVR236 FPTDQLTPDQ246 ERSLMFMQWG256 QLLDHDLDFT266 PEPAARASFV 276 TGVNCETSCV286 QQPPCFPLKI296 PPNDPRIKNQ306 ADCIPFFRSP317 ACPGSNITIR 327 NQINALTSFV337 DASMVYGSEE347 PLARNLRNMS357 NQLGLLAVNQ367 RFQDNGRALL 377 PFDNLHDDPC387 LLTNRSARIP397 CFLAGDTRSS407 EMPELTSMHT417 LLLREHNRLA 427 TELKSLNPRW437 DGERLYQEAR447 KIVGAMVQII457 TYRDYLPLVL467 GPTAMRKYLP 477 TYRSYNDSVD487 PRIANVFTNA497 FRYGHTLIQP507 FMFRLDNRYQ517 PMEPNPRVPL 527 SRVFFASWRV537 VLEGGIDPIL547 RGLMATPAKL557 NRQNQIAVDE567 IRERLFEQVM 577 RIGLDLPALN587 MQRSRDHGLP597 GYNAWRRFCG607 LPQPETVGQL617 GTVLRNLKLA 627 RKLMEQYGTP637 NNIDIWMGGV647 SEPLKRKGRV657 GPLLACIIGT667 QFRKLRDGDR 677 FWWENEGVFS687 MQQRQALAQI697 SLPRIICDNT707 GITTVSKNNI717 FMSNSYPRDF 727 VNCSTLPALN737 LASWRE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Drug metabolism - other enzymes | hsa00983 | Affiliated Target |
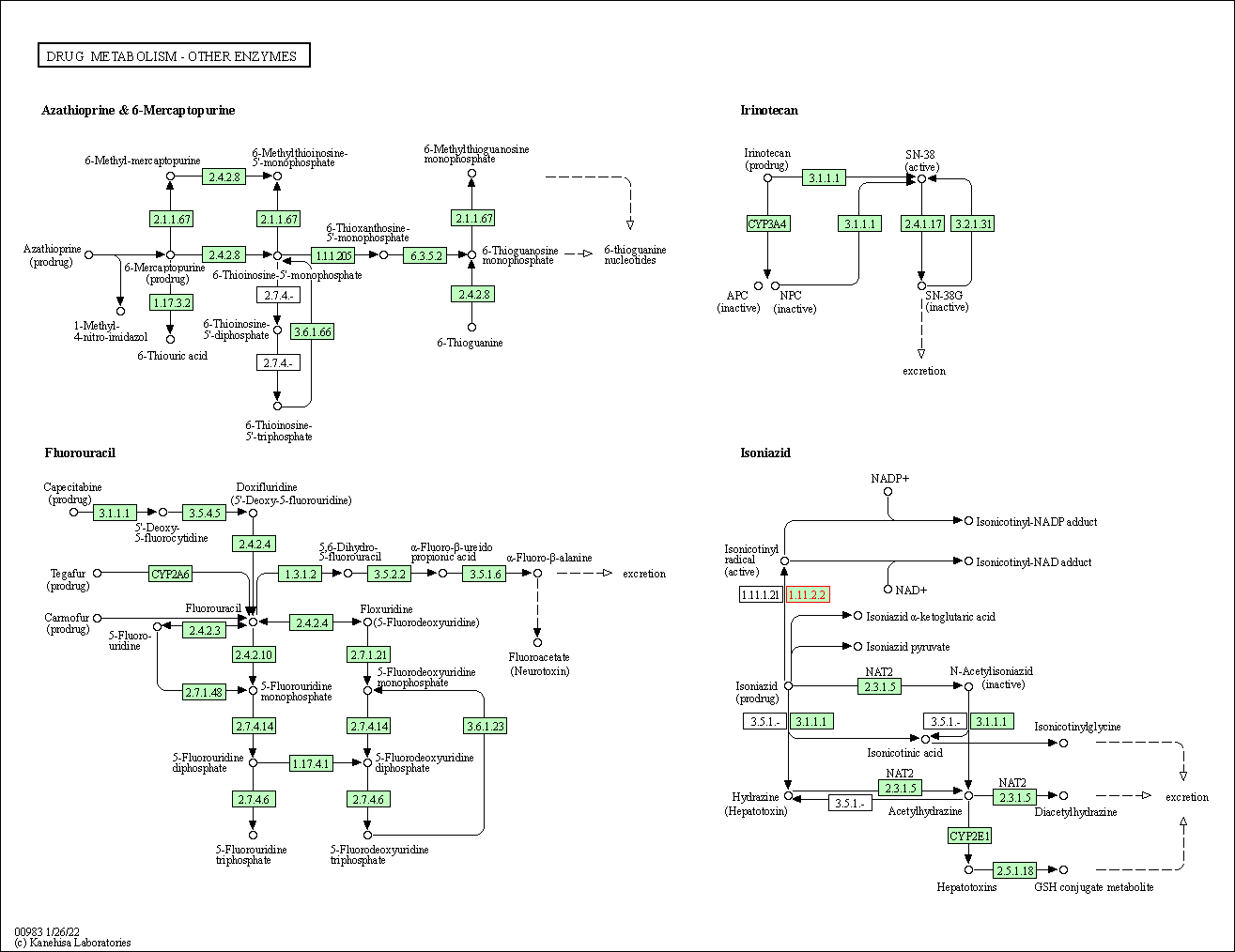
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Phagosome | hsa04145 | Affiliated Target |
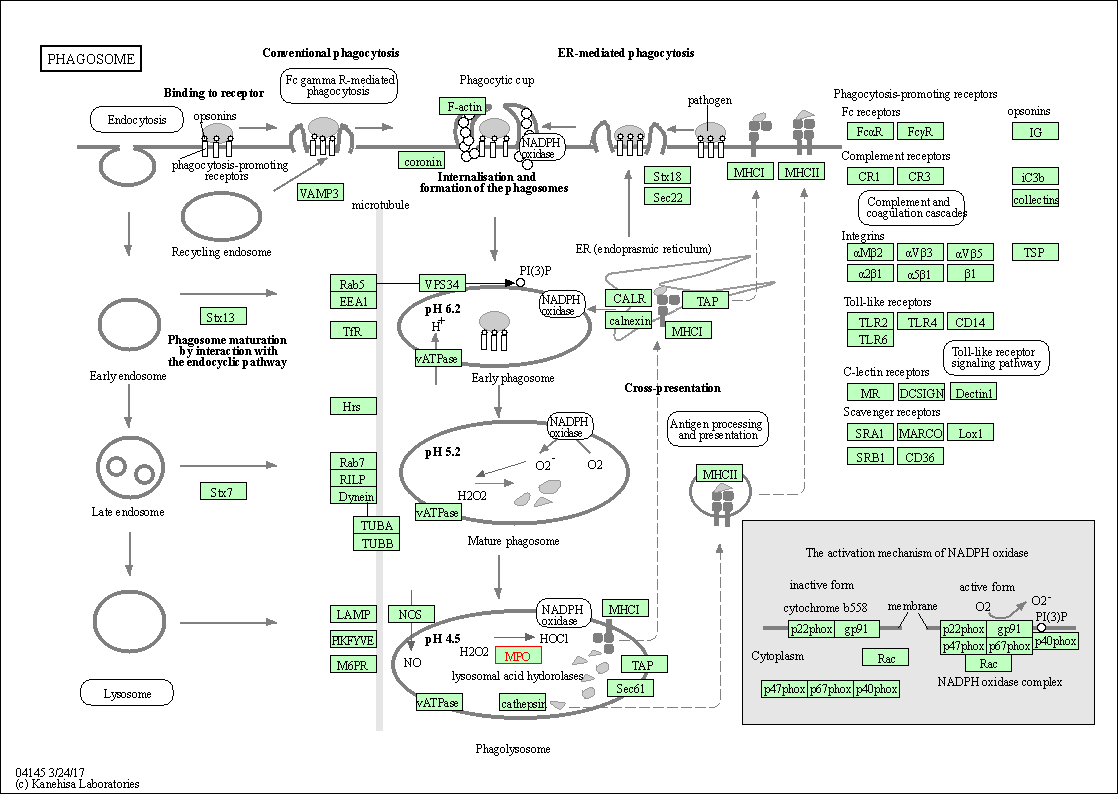
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Neutrophil extracellular trap formation | hsa04613 | Affiliated Target |
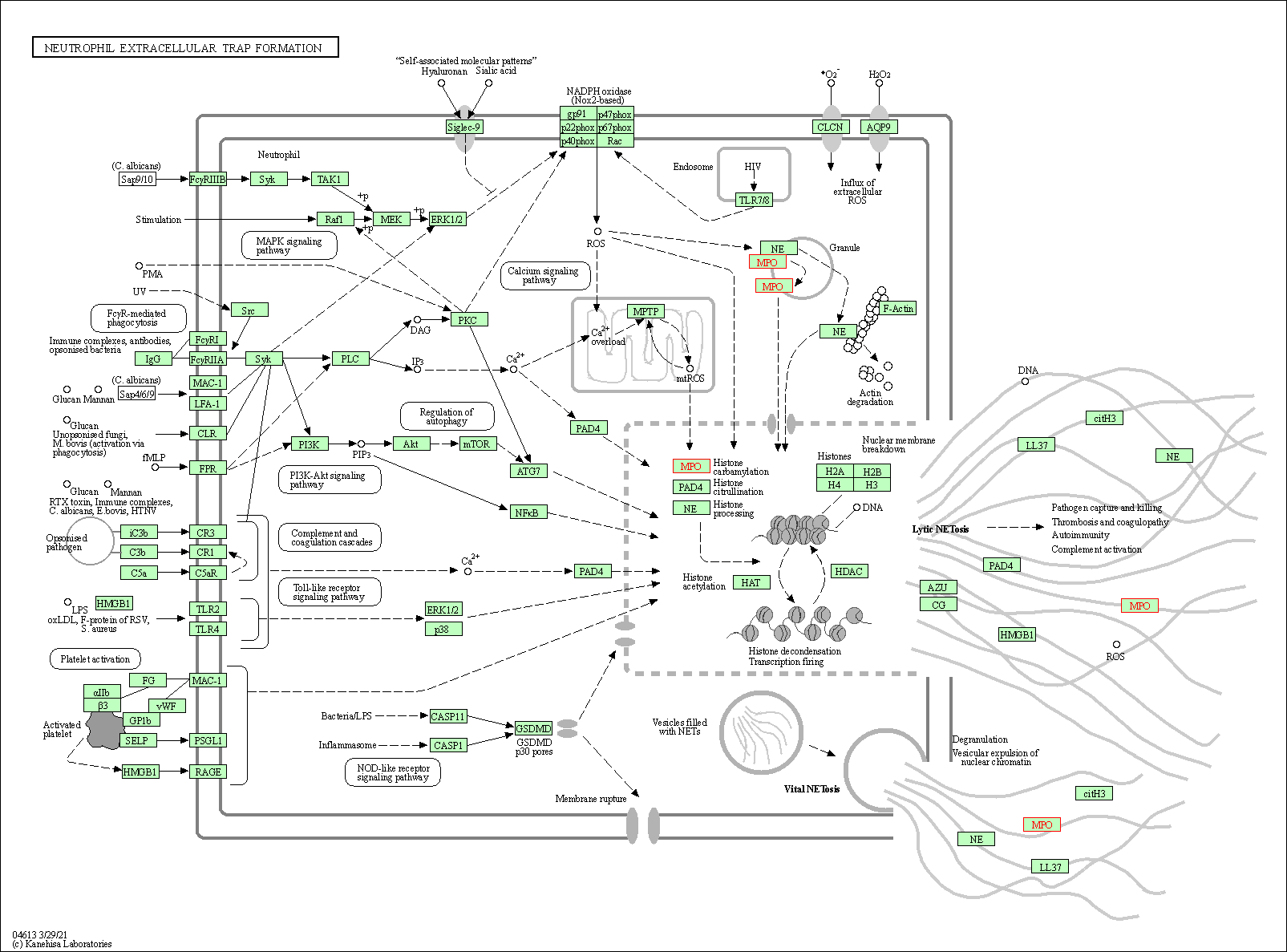
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 2.45E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.77E-01 | Radiality | 1.29E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 1.15E+01 | Topological coefficient | 2.63E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 2 KEGG Pathways | + | ||||
| 1 | Phagosome | |||||
| 2 | Transcriptional misregulation in cancer | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | C-MYB transcription factor network | |||||
| 2 | IL23-mediated signaling events | |||||
| WikiPathways | [+] 3 WikiPathways | + | ||||
| 1 | Folate Metabolism | |||||
| 2 | Vitamin B12 Metabolism | |||||
| 3 | Selenium Micronutrient Network | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | In vitro antibacterial activity of E-101 Solution, a novel myeloperoxidase-mediated antimicrobial, against Gram-positive and Gram-negative pathogens. J Antimicrob Chemother. 2011 Feb;66(2):335-42. | |||||
| REF 2 | ClinicalTrials.gov (NCT01297959) Efficacy and Safety of E-101 Solution for Preventing Surgical Site Infections After Colorectal Surgery. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT04986202) A Randomised, Double-blind, Placebo-controlled, Multi-center Sequential Phase 2b and Phase 3 Study to Evaluate the Efficacy and Safety of AZD4831 Administered for Up to 48 Weeks in Participants With Heart Failure With Left Ventricular Ejection Fraction > 40%. U.S.National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT02388295) AZD3241 PET MSA Trial, Phase 2, Randomized,12 Week Safety and Tolerability Trial With PET in MSA Patients. U.S. National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 6 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7728). | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800024670) | |||||
| REF 8 | Effect of the myeloperoxidase inhibitor AZD3241 on microglia: a PET study in Parkinson's disease. Brain. 2015 Sep;138(Pt 9):2687-700. | |||||
| REF 9 | Myeloperoxidase expression as a potential determinant of parthenolide-induced apoptosis in leukemia bulk and leukemia stem cells. J Pharmacol Exp Ther. 2010 Nov;335(2):389-400. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2789). | |||||
| REF 11 | Native glycosylation and binding of the antidepressant paroxetine in a low-resolution crystal structure of human myeloperoxidase. Acta Crystallogr D Struct Biol. 2022 Sep 1;78(Pt 9):1099-1109. | |||||
| REF 12 | A structurally dynamic N-terminal region drives function of the staphylococcal peroxidase inhibitor (SPIN). J Biol Chem. 2018 Feb 16;293(7):2260-2271. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

