Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T25608
(Former ID: TTDNC00653)
|
|||||
| Target Name |
Free fatty acid receptor 1 (GPR40)
|
|||||
| Synonyms |
Gprotein coupled receptor 40; FFAR1
Click to Show/Hide
|
|||||
| Gene Name |
FFAR1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Type 2 diabetes mellitus [ICD-11: 5A11] | |||||
| Function |
G-protein coupled receptor for medium and long chain saturated and unsaturated fatty acids that plays an important role in glucose homeostasis. Fatty acid binding increases glucose- stimulated insulin secretion, and may also enhance the secretion of glucagon-like peptide 1 (GLP-1). May also play a role in bone homeostasis; receptor signaling activates pathways that inhibit osteoclast differentiation. Ligand binding leads to a conformation change that triggers signaling via G-proteins that activate phospholipase C, leading to an increase of the intracellular calcium concentration. Seems to act through a G(q) and G(i)-mediated pathway.
Click to Show/Hide
|
|||||
| BioChemical Class |
GPCR rhodopsin
|
|||||
| UniProt ID | ||||||
| Sequence |
MDLPPQLSFGLYVAAFALGFPLNVLAIRGATAHARLRLTPSLVYALNLGCSDLLLTVSLP
LKAVEALASGAWPLPASLCPVFAVAHFFPLYAGGGFLAALSAGRYLGAAFPLGYQAFRRP CYSWGVCAAIWALVLCHLGLVFGLEAPGGWLDHSNTSLGINTPVNGSPVCLEAWDPASAG PARFSLSLLLFFLPLAITAFCYVGCLRALARSGLTHRRKLRAAWVAGGALLTLLLCVGPY NASNVASFLYPNLGGSWRKLGLITGAWSVVLNPLVTGYLGRGPGLKTVCAARTQGGKSQK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T88MHO | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 10 Clinical Trial Drugs | + | ||||
| 1 | Fasiglifam hemihydrate | Drug Info | Phase 3 | Type-2 diabetes | [2] | |
| 2 | TAK-875 | Drug Info | Phase 3 | Type-2 diabetes | [3], [4] | |
| 3 | JTT-851 | Drug Info | Phase 2 | Type-2 diabetes | [5], [6] | |
| 4 | P-1736 | Drug Info | Phase 2 | Diabetic complication | [7] | |
| 5 | PBI-4050 | Drug Info | Phase 2 | Idiopathic pulmonary fibrosis | [8] | |
| 6 | CPL207-280CA | Drug Info | Phase 1 | Type 2 diabetes | [9] | |
| 7 | LY-2881835 | Drug Info | Phase 1 | Type-2 diabetes | [10] | |
| 8 | LY2922470 | Drug Info | Phase 1 | Type 2 diabetes | [11] | |
| 9 | P11187 | Drug Info | Phase 1 | Type-2 diabetes | [12] | |
| 10 | SHR0534 | Drug Info | Phase 1 | Type 2 diabetes | [13] | |
| Preclinical Drug(s) | [+] 3 Preclinical Drugs | + | ||||
| 1 | BMS-986118 | Drug Info | Preclinical | Type 2 diabetes | [14] | |
| 2 | CNX-011-67 | Drug Info | Preclinical | Type 2 diabetes | [9] | |
| 3 | DS-1558 | Drug Info | Preclinical | Type 2 diabetes | [9] | |
| Mode of Action | [+] 4 Modes of Action | + | ||||
| Modulator | [+] 5 Modulator drugs | + | ||||
| 1 | Fasiglifam hemihydrate | Drug Info | [1], [15] | |||
| 2 | TAK-875 | Drug Info | [16] | |||
| 3 | P-1736 | Drug Info | [7] | |||
| 4 | LY-2881835 | Drug Info | [17] | |||
| 5 | P11187 | Drug Info | [17] | |||
| Agonist | [+] 16 Agonist drugs | + | ||||
| 1 | JTT-851 | Drug Info | [17] | |||
| 2 | PBI-4050 | Drug Info | [18] | |||
| 3 | CPL207-280CA | Drug Info | [9] | |||
| 4 | LY2922470 | Drug Info | [19] | |||
| 5 | SHR0534 | Drug Info | [9] | |||
| 6 | BMS-986118 | Drug Info | [20] | |||
| 7 | DS-1558 | Drug Info | [22] | |||
| 8 | alpha-linolenic acid | Drug Info | [23] | |||
| 9 | AMG-837 | Drug Info | [24] | |||
| 10 | CNX-011 | Drug Info | [25] | |||
| 11 | GW9508 | Drug Info | [26] | |||
| 12 | medica 16 | Drug Info | [27] | |||
| 13 | PMID18477808CB | Drug Info | [28] | |||
| 14 | TUG-424 | Drug Info | [29] | |||
| 15 | TUG-469 | Drug Info | [25] | |||
| 16 | TUG-770 | Drug Info | [30] | |||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | CNX-011-67 | Drug Info | [21] | |||
| Antagonist | [+] 1 Antagonist drugs | + | ||||
| 1 | GW1100 | Drug Info | [26] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Insulin secretion | hsa04911 | Affiliated Target |
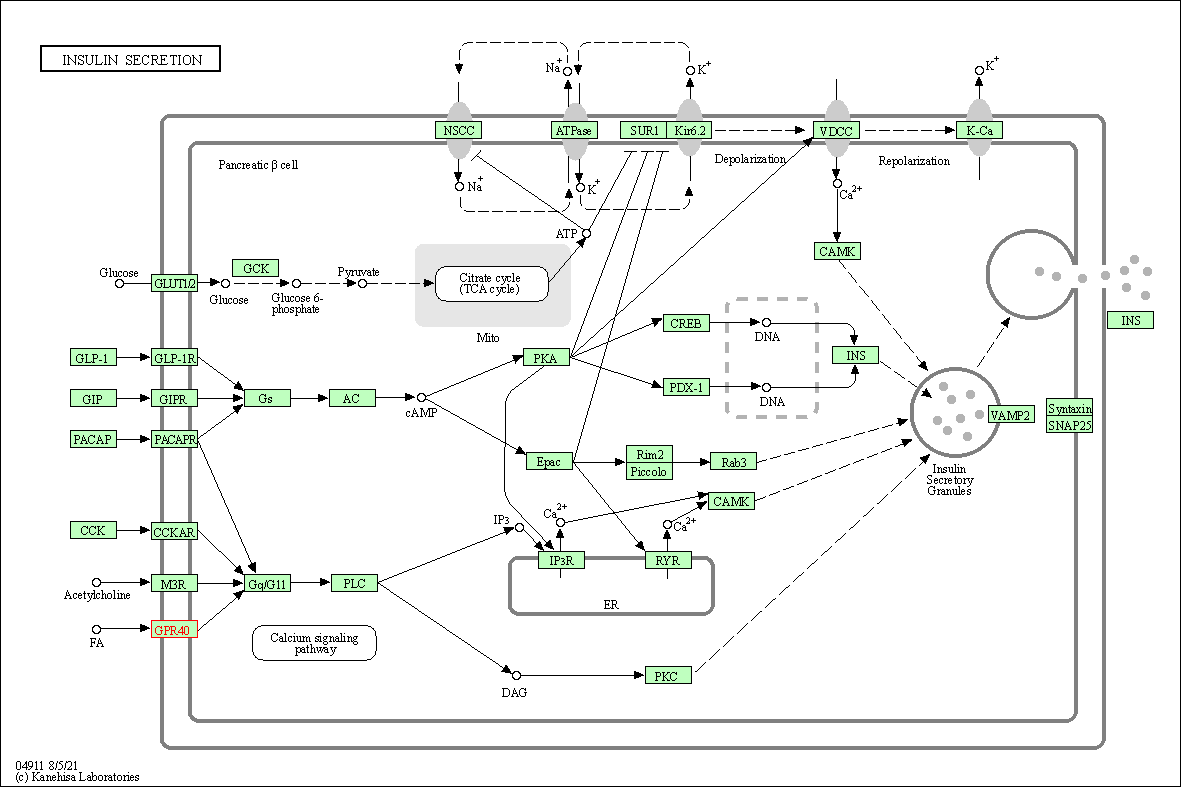
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 1.42E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.87E-01 | Radiality | 1.32E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 2.85E+01 | Topological coefficient | 5.00E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Drug Resistance Mutation (DRM) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Insulin secretion | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | G alpha (q) signalling events | |||||
| 2 | Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion | |||||
| WikiPathways | [+] 6 WikiPathways | + | ||||
| 1 | GPCRs, Class A Rhodopsin-like | |||||
| 2 | Incretin Synthesis, Secretion, and Inactivation | |||||
| 3 | Gastrin-CREB signalling pathway via PKC and MAPK | |||||
| 4 | Integration of energy metabolism | |||||
| 5 | GPCR ligand binding | |||||
| 6 | GPCR downstream signaling | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | A novel antidiabetic drug, fasiglifam/TAK-875, acts as an ago-allosteric modulator of FFAR1. PLoS One. 2013 Oct 10;8(10):e76280. | |||||
| REF 2 | ClinicalTrials.gov (NCT02015780) Fasiglifam in Type 2 Diabetic Subjects With Chronic Kidney Disease Stage 4 or 5 on Hemodialysis. U.S. National Institutes of Health. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6484). | |||||
| REF 4 | ClinicalTrials.gov (NCT01609582) Study of TAK-875 in Adults With Type 2 Diabetes and Cardiovascular Disease or Risk Factors for Cardiovascular Disease. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT01699737) Safety and Efficacy Study of JTT-851 in Patients With Type 2 Diabetes Mellitus. U.S. National Institutes of Health. | |||||
| REF 6 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 7 | Discovery of p1736, a novel antidiabetic compound that improves peripheral insulin sensitivity in mice models. PLoS One. 2013 Oct 23;8(10):e77946. | |||||
| REF 8 | Clinical pipeline report, company report or official report of Liminal BioSciences. | |||||
| REF 9 | Targeting metabolic dysregulation for fibrosis therapy. Nat Rev Drug Discov. 2020 Jan;19(1):57-75. | |||||
| REF 10 | ClinicalTrials.gov (NCT01358981) A Study of LY2881835 in Healthy People and People With Diabetes. U.S. National Institutes of Health. | |||||
| REF 11 | ClinicalTrials.gov (NCT01867216) A Study of Multiple Doses of LY2922470 in Participants With Diabetes. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT01874366) Determination of Safety,Tolerability,Pharmacokinetics,Food Effect& Pharmacodynamics of Single & Multiple Doses of P11187. U.S. National Institutes of Health. | |||||
| REF 13 | ClinicalTrials.gov (NCT03006159) A Phase 1, Randomized, Placebo-controlled, Multiple Dose Escalation Study to Investigate Safety, Pharmacokinetics, and Pharmacodynamics of SHR0534 in Chinese Type 2 Diabetic Patients. U.S. National Institutes of Health. | |||||
| REF 14 | Free fatty acid receptor agonists for the treatment of type 2 diabetes: drugs in preclinical to phase II clinical development. Expert Opin Investig Drugs. 2016 Aug;25(8):871-90. | |||||
| REF 15 | TAK-875, a GPR40/FFAR1 agonist, in combination with metformin prevents progression of diabetes and beta-cell dysfunction in Zucker diabetic fatty rats. Br J Pharmacol. 2013 Oct;170(3):568-80. | |||||
| REF 16 | Efficacy and safety of fasiglifam (TAK-875), a G protein-coupled receptor 40 agonist, in Japanese patients with type 2 diabetes inadequately controlled by diet and exercise: a randomized, double-blind, placebo-controlled, phase III trial. Diabetes Obes Metab. 2015 Jul;17(7):675-81. | |||||
| REF 17 | Treatment of Type 2 Diabetes by Free Fatty Acid Receptor Agonists. Front Endocrinol (Lausanne) 2014; 5: 137. | |||||
| REF 18 | Phase 2 clinical trial of PBI-4050 in patients with idiopathic pulmonary fibrosis. Eur Respir J. 2019 Mar 18;53(3):1800663. | |||||
| REF 19 | The Discovery, Preclinical, and Early Clinical Development of Potent and Selective GPR40 Agonists for the Treatment of Type 2 Diabetes Mellitus (LY2881835, LY2922083, and LY2922470). J Med Chem. 2016 Dec 22;59(24):10891-10916. | |||||
| REF 20 | Discovery of Potent and Orally Bioavailable Dihydropyrazole GPR40 Agonists. J Med Chem. 2018 Feb 8;61(3):681-694. | |||||
| REF 21 | CNX-011-67, a novel GPR40 agonist, enhances glucose responsiveness, insulin secretion and islet insulin content in n-STZ rats and in islets from type 2 diabetic patients. BMC Pharmacol Toxicol. 2014 Mar 25;15:19. | |||||
| REF 22 | Discovery of DS-1558: A Potent and Orally Bioavailable GPR40 Agonist. ACS Med Chem Lett. 2015 Jan 13;6(3):266-70. | |||||
| REF 23 | Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003 Mar 13;422(6928):173-6. | |||||
| REF 24 | Identification and pharmacological characterization of multiple allosteric binding sites on the free fatty acid 1 receptor. Mol Pharmacol. 2012 Nov;82(5):843-59. | |||||
| REF 25 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 225). | |||||
| REF 26 | Pharmacological regulation of insulin secretion in MIN6 cells through the fatty acid receptor GPR40: identification of agonist and antagonist small... Br J Pharmacol. 2006 Jul;148(5):619-28. | |||||
| REF 27 | A human cell surface receptor activated by free fatty acids and thiazolidinedione drugs. Biochem Biophys Res Commun. 2003 Feb 7;301(2):406-10. | |||||
| REF 28 | Selective small-molecule agonists of G protein-coupled receptor 40 promote glucose-dependent insulin secretion and reduce blood glucose in mice. Diabetes. 2008 Aug;57(8):2211-9. | |||||
| REF 29 | Discovery of potent and selective agonists for the free fatty acid receptor 1 (FFA(1)/GPR40), a potential target for the treatment of type II diabetes. J Med Chem. 2008 Nov 27;51(22):7061-4. | |||||
| REF 30 | Discovery of TUG-770: A Highly Potent Free Fatty Acid Receptor 1 (FFA1/GPR40) Agonist for Treatment of Type 2 Diabetes. ACS Med Chem Lett. 2013 May 9;4(5):441-445. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

