Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T34296
(Former ID: TTDC00150)
|
|||||
| Target Name |
Matrix metalloproteinase-13 (MMP-13)
|
|||||
| Synonyms |
Matrix metalloproteinase 13; Collagenase-3; Collagenase 3
Click to Show/Hide
|
|||||
| Gene Name |
MMP13
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Cleaves triple helical collagens, including type I, type II and type III collagen, but has the highest activity with soluble type II collagen. Can also degrade collagen type IV, type XIV and type X. May also function by activating or degrading key regulatory proteins, such as TGFB1 and CTGF. Plays a role in wound healing, tissue remodeling, cartilage degradation, bone development, bone mineralization and ossification. Required for normal embryonic bone development and ossification. Plays a role in the healing of bone fractures via endochondral ossification. Plays a role in wound healing, probably by a mechanism that involves proteolytic activation of TGFB1 and degradation of CTGF. Plays a role in keratinocyte migration during wound healing. May play a role in cell migration and in tumor cell invasion. Plays a role in the degradation of extracellular matrix proteins including fibrillar collagen, fibronectin, TNC and ACAN.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.24.-
|
|||||
| Sequence |
MHPGVLAAFLFLSWTHCRALPLPSGGDEDDLSEEDLQFAERYLRSYYHPTNLAGILKENA
ASSMTERLREMQSFFGLEVTGKLDDNTLDVMKKPRCGVPDVGEYNVFPRTLKWSKMNLTY RIVNYTPDMTHSEVEKAFKKAFKVWSDVTPLNFTRLHDGIADIMISFGIKEHGDFYPFDG PSGLLAHAFPPGPNYGGDAHFDDDETWTSSSKGYNLFLVAAHEFGHSLGLDHSKDPGALM FPIYTYTGKSHFMLPDDDVQGIQSLYGPGDEDPNPKHPKTPDKCDPSLSLDAITSLRGET MIFKDRFFWRLHPQQVDAELFLTKSFWPELPNRIDAAYEHPSHDLIFIFRGRKFWALNGY DILEGYPKKISELGLPKEVKKISAAVHFEDTGKTLLFSGNQVWRYDDTNHIMDKDYPRLI EEDFPGIGDKVDAVYEKNGYIYFFNGPIQFEYSIWSNRIVRVMPANSILWC Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T62P0D | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | Curcumin | Drug Info | Phase 3 | Solid tumour/cancer | [2], [3] | |
| 2 | Apratastat | Drug Info | Phase 2 | Rheumatoid arthritis | [4], [5] | |
| 3 | PMID17935984C1 | Drug Info | Clinical trial | Pain | [6], [7] | |
| Discontinued Drug(s) | [+] 2 Discontinued Drugs | + | ||||
| 1 | GM6001 | Drug Info | Discontinued in Phase 2 | Corneal ulcer | [8] | |
| 2 | RS-130830 | Drug Info | Discontinued in Phase 2 | Hepatitis C virus infection | [9] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 43 Inhibitor drugs | + | ||||
| 1 | Curcumin | Drug Info | [1] | |||
| 2 | PMID17935984C1 | Drug Info | [6] | |||
| 3 | PMID29130358-Compound-Figure16(9a) | Drug Info | [13] | |||
| 4 | PMID29130358-Compound-Figure16(9b) | Drug Info | [13] | |||
| 5 | PMID29130358-Compound-Figure16(9c) | Drug Info | [13] | |||
| 6 | PMID29130358-Compound-Figure18(14) | Drug Info | [13] | |||
| 7 | GM6001 | Drug Info | [14] | |||
| 8 | RS-130830 | Drug Info | [15] | |||
| 9 | (+/-)5-(biphenyl-4-yl)-3-hydroxypentanoic acid | Drug Info | [16] | |||
| 10 | 1-(4-Methoxy-benzenesulfonyl)-heptane-3-thiol | Drug Info | [17] | |||
| 11 | 1-Methyloxy-4-Sulfone-Benzene | Drug Info | [18] | |||
| 12 | 2-(2-(biphenyl-4-yl)ethylsulfonyl)acetic acid | Drug Info | [16] | |||
| 13 | 2-(4-methoxybenzylthio)-6-methylpyrimidin-4-ol | Drug Info | [19] | |||
| 14 | 2-(biphenyl-4-ylsulfonamido)pentanedioic acid | Drug Info | [19] | |||
| 15 | 2-(Biphenyl-4-ylsulfonyl)N-hydroxybenzamide | Drug Info | [20] | |||
| 16 | 3-(4-Methoxy-benzenesulfonyl)-cyclohexanethiol | Drug Info | [21] | |||
| 17 | 3-(4-Methoxy-benzenesulfonyl)-cyclopentanethiol | Drug Info | [21] | |||
| 18 | 3-(4-Methoxy-benzenesulfonyl)-hexane-1-thiol | Drug Info | [17] | |||
| 19 | 3-(4-Methoxy-benzenesulfonyl)-pentane-1-thiol | Drug Info | [17] | |||
| 20 | 3-(4-Methoxy-benzenesulfonyl)-propane-1-thiol | Drug Info | [17] | |||
| 21 | 3-(4-Phenoxy-benzenesulfonyl)-cyclohexanethiol | Drug Info | [21] | |||
| 22 | 3-(4-Phenoxy-benzenesulfonyl)-propane-1-thiol | Drug Info | [17] | |||
| 23 | 3-Benzenesulfonyl-heptanoic acid hydroxyamide | Drug Info | [22] | |||
| 24 | 3-Cyclohexanesulfonyl-heptanoic acid hydroxyamide | Drug Info | [22] | |||
| 25 | 3-Methylpyridine | Drug Info | [18] | |||
| 26 | 4-(2,2'-bithiophen-5-ylmethyleneamino)phenol | Drug Info | [1] | |||
| 27 | 4-(methyl(4-phenylthiazol-2-yl)amino)phenol | Drug Info | [1] | |||
| 28 | 5-(4'-cyanobiphenyl-4-yl)-3-hydroxypentanoic acid | Drug Info | [16] | |||
| 29 | Ethyl 2-cyano-2-(quinoxalin-2(1H)-ylidene)acetate | Drug Info | [1] | |||
| 30 | Hydroxyaminovaline | Drug Info | [18] | |||
| 31 | MMI270 | Drug Info | [23] | |||
| 32 | N-Hydroxy-2-(4-phenoxy-benzenesulfonyl)benzamide | Drug Info | [20] | |||
| 33 | N1,N3-bis(3-methoxybenzyl)isophthalamide | Drug Info | [24] | |||
| 34 | N4,N6-dibenzylpyrimidine-4,6-dicarboxamide | Drug Info | [25] | |||
| 35 | PD-156 | Drug Info | [26], [27] | |||
| 36 | PKF-242-484 | Drug Info | [28] | |||
| 37 | PMID22017539C15 | Drug Info | [29] | |||
| 38 | SL422 | Drug Info | [30] | |||
| 39 | SR-973 | Drug Info | [31] | |||
| 40 | UK-356618 | Drug Info | [32] | |||
| 41 | WAY-151693 | Drug Info | [18] | |||
| 42 | WAY170523 | Drug Info | [33] | |||
| 43 | [2-(Biphenyl-4-sulfonyl)phenyl]acetic Acid | Drug Info | [20] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Apratastat | Drug Info | [10], [11], [12] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Acetohydroxamic acid | Ligand Info | |||||
| Structure Description | Crystal structure analysis of the MMP13 catalytic domain in complex with specific inhibitor | PDB:2OW9 | ||||
| Method | X-ray diffraction | Resolution | 1.74 Å | Mutation | No | [14] |
| PDB Sequence |
YNVFPRTLKW
92 SKMNLTYRIV102 NYTPDMTHSE112 VEKAFKKAFK122 VWSDVTPLNF132 TRLHDGIADI 142 MISFGIKEHG152 DFYPFDGPSG162 LLAHAFPPGP172 NYGGDAHFDD182 DETWTSSSKG 192 YNLFLVAAHE202 FGHSLGLDHS212 KDPGALMFPI222 YTYTGKSHFM232 LPDDDVQGIQ 242 SLYGPGD
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: PMID17935984C1 | Ligand Info | |||||
| Structure Description | Crystal structure of the catalytic domain of MMP-13 complexed with a potent pyrimidinetrione inhibitor | PDB:1YOU | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | No | [34] |
| PDB Sequence |
YNVFPRTLKW
113 SKMNLTYRIV123 NYTPDMTHSE133 VEKAFKKAFK143 VWSDVTPLNF153 TRLHDGIADI 163 MISFGIKEHG173 DFYPFDGPSG183 LLAHAFPPGP193 NYGGDAHFDD203 DETWTSSSKG 213 YNLFLVAAHE223 FGHSLGLDHS233 KDPGALMFPI243 YTYTGKSHFM253 LPDDDVQGIQ 263 SLYGPGDE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| IL-17 signaling pathway | hsa04657 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |
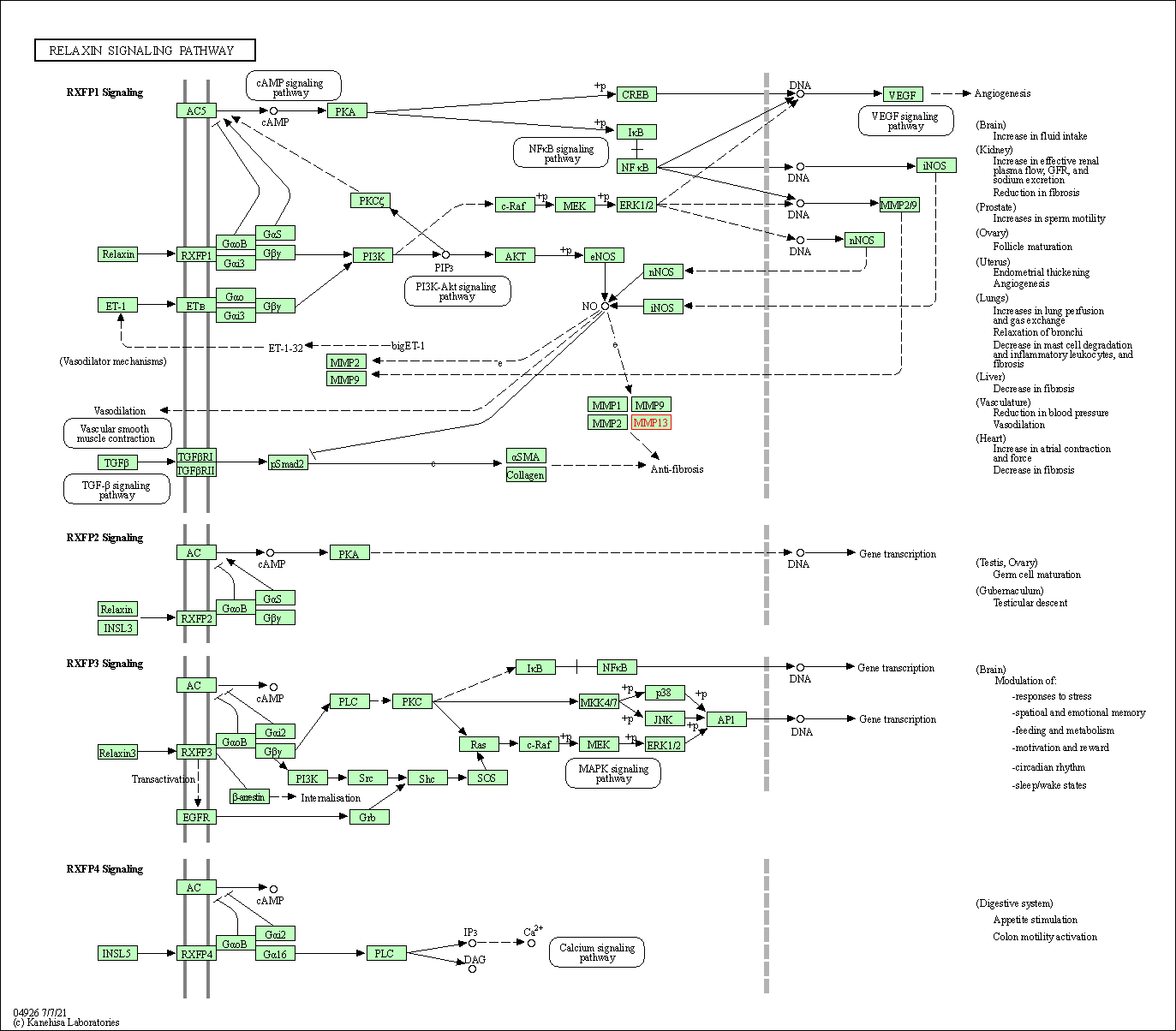
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 3 | Degree centrality | 3.22E-04 | Betweenness centrality | 9.49E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.03E-01 | Radiality | 1.35E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 2.10E+01 | Topological coefficient | 3.45E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | IL1 Signaling Pathway | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Alzheimer disease-presenilin pathway | |||||
| 2 | Plasminogen activating cascade | |||||
| PID Pathway | [+] 1 PID Pathways | + | ||||
| 1 | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling | |||||
| Reactome | [+] 4 Reactome Pathways | + | ||||
| 1 | Collagen degradation | |||||
| 2 | Degradation of the extracellular matrix | |||||
| 3 | Activation of Matrix Metalloproteinases | |||||
| 4 | Assembly of collagen fibrils and other multimeric structures | |||||
| WikiPathways | [+] 5 WikiPathways | + | ||||
| 1 | Endochondral Ossification | |||||
| 2 | Activation of Matrix Metalloproteinases | |||||
| 3 | Oncostatin M Signaling Pathway | |||||
| 4 | AGE/RAGE pathway | |||||
| 5 | Matrix Metalloproteinases | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| Target QSAR Model | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | High throughput screening of potentially selective MMP-13 exosite inhibitors utilizing a triple-helical FRET substrate. Bioorg Med Chem. 2009 Feb 1;17(3):990-1005. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7000). | |||||
| REF 3 | Nanocurcumin: a promising therapeutic advancement over native curcumin. Crit Rev Ther Drug Carrier Syst. 2013;30(4):331-68. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6482). | |||||
| REF 5 | ClinicalTrials.gov (NCT00095342) Study Evaluating TMI-005 in Active Rheumatoid Arthritis. U.S. National Institutes of Health. | |||||
| REF 6 | Potent, selective spiropyrrolidine pyrimidinetrione inhibitors of MMP-13. Bioorg Med Chem Lett. 2007 Dec 1;17(23):6529-34. | |||||
| REF 7 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6526). | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800001387) | |||||
| REF 9 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010620) | |||||
| REF 10 | Acetylenic TACE inhibitors. Part 3: Thiomorpholine sulfonamide hydroxamates. Bioorg Med Chem Lett. 2006 Mar 15;16(6):1605-9. | |||||
| REF 11 | Drug insight: tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis. Nat Clin Pract Rheumatol. 2008 Jun;4(6):300-9. | |||||
| REF 12 | Drug evaluation: apratastat, a novel TACE/MMP inhibitor for rheumatoid arthritis. Curr Opin Investig Drugs. 2006 Nov;7(11):1014-9. | |||||
| REF 13 | Gelatinase inhibitors: a patent review (2011-2017).Expert Opin Ther Pat. 2018 Jan;28(1):31-46. | |||||
| REF 14 | Discovery and characterization of a novel inhibitor of matrix metalloprotease-13 that reduces cartilage damage in vivo without joint fibroplasia si... J Biol Chem. 2007 Sep 21;282(38):27781-91. | |||||
| REF 15 | Structure-based design of potent and selective inhibitors of collagenase-3 (MMP-13). Bioorg Med Chem Lett. 2005 Feb 15;15(4):1101-6. | |||||
| REF 16 | The identification of beta-hydroxy carboxylic acids as selective MMP-12 inhibitors. Bioorg Med Chem Lett. 2009 Oct 1;19(19):5760-3. | |||||
| REF 17 | Discovery of a novel series of selective MMP inhibitors: identification of the gamma-sulfone-thiols. Bioorg Med Chem Lett. 1999 Apr 5;9(7):943-8. | |||||
| REF 18 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 19 | Ranking the selectivity of PubChem screening hits by activity-based protein profiling: MMP13 as a case study. Bioorg Med Chem. 2009 Feb 1;17(3):1101-8. | |||||
| REF 20 | Design, synthesis, biological evaluation, and NMR studies of a new series of arylsulfones as selective and potent matrix metalloproteinase-12 inhib... J Med Chem. 2009 Oct 22;52(20):6347-61. | |||||
| REF 21 | Synthesis and identification of conformationally constrained selective MMP inhibitors. Bioorg Med Chem Lett. 1999 Jul 5;9(13):1757-60. | |||||
| REF 22 | Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J Med Chem. 2000 Jun 15;43(12):2324-31. | |||||
| REF 23 | Heterocycle-based MMP inhibitors with P2' substituents. Bioorg Med Chem Lett. 2001 Apr 23;11(8):1009-13. | |||||
| REF 24 | Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment ... Bioorg Med Chem Lett. 2010 Jan 15;20(2):576-80. | |||||
| REF 25 | Calculation of binding free energies for non-zinc chelating pyrimidine dicarboxamide inhibitors with MMP-13. Bioorg Med Chem Lett. 2009 Jan 1;19(1):47-50. | |||||
| REF 26 | Selective matrix metalloproteinase inhibition attenuates progression of left ventricular dysfunction and remodeling in dogs with chronic heart fail... Am J Physiol Heart Circ Physiol. 2006 Jun;290(6):H2522-7. | |||||
| REF 27 | Effects of selective matrix metalloproteinase inhibitor (PG-116800) to prevent ventricular remodeling after myocardial infarction: results of the P... J Am Coll Cardiol. 2006 Jul 4;48(1):15-20. | |||||
| REF 28 | A cassette-dosing approach for improvement of oral bioavailability of dual TACE/MMP inhibitors. Bioorg Med Chem Lett. 2006 May 15;16(10):2632-6. | |||||
| REF 29 | Fragment-based discovery of indole inhibitors of matrix metalloproteinase-13. J Med Chem. 2011 Dec 8;54(23):8174-87. | |||||
| REF 30 | Design, synthesis, and structure-activity relationships of macrocyclic hydroxamic acids that inhibit tumor necrosis factor alpha release in vitro and in vivo. J Med Chem. 2001 Aug 2;44(16):2636-60. | |||||
| REF 31 | Synthesis and evaluation of succinoyl-caprolactam gamma-secretase inhibitors. Bioorg Med Chem Lett. 2006 May 1;16(9):2357-63. | |||||
| REF 32 | A potent, selective inhibitor of matrix metalloproteinase-3 for the topical treatment of chronic dermal ulcers. J Med Chem. 2003 Jul 31;46(16):3514-25. | |||||
| REF 33 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1637). | |||||
| REF 34 | Potent pyrimidinetrione-based inhibitors of MMP-13 with enhanced selectivity over MMP-14. Bioorg Med Chem Lett. 2005 Apr 1;15(7):1807-10. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

