Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T37848
(Former ID: TTDS00418)
|
|||||
| Target Name |
Albendazole monooxygenase (CYP3A4)
|
|||||
| Synonyms |
Taurochenodeoxycholate 6-alpha-hydroxylase; Quinine 3-monooxygenase; P450-PCN1; Nifedipine oxidase; NF-25; HLp; Cytochrome P450-PCN1; Cytochrome P450 NF-25; Cytochrome P450 HLp; Cytochrome P450 3A4; Cytochrome P450 3A3; CYPIIIA4; CYPIIIA3; CYP3A3; Albendazole sulfoxidase; Albendazole monooxygenase (sulfoxide-forming); 1,8-cineole 2-exo-monooxygenase
Click to Show/Hide
|
|||||
| Gene Name |
CYP3A4
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e. g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide. Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro. Catalyzes sulfoxidation of the anthelmintics albendazole and fenbendazole. Cytochromes P450 are a group of heme-thiolate monooxygenases.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.14.-
|
|||||
| Sequence |
MALIPDLAMETWLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMF
DMECHKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISI AEDEEWKRLRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYS MDVITSTSFGVNIDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICV FPREVTNFLRKSVKRMKESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSI IFIFAGYETTSSVLSFIMYELATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVV NETLRLFPIAMRLERVCKKDVEINGMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFS KKNKDNIDPYIYTPFGSGPRNCIGMRFALMNMKLALIRVLQNFSFKPCKETQIPLKLSLG GLLQPEKPVVLKVESRDGTVSGA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A00232 ; BADD_A00376 ; BADD_A00541 ; BADD_A00659 ; BADD_A01336 ; BADD_A01965 ; BADD_A02053 ; BADD_A02189 ; BADD_A02216 ; BADD_A02978 ; BADD_A03877 ; BADD_A04155 ; BADD_A04220 ; BADD_A04253 ; BADD_A04263 ; BADD_A04424 ; BADD_A05280 ; BADD_A05957 ; BADD_A05962 ; BADD_A06627 | |||||
| HIT2.0 ID | T85AXP | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | Curcumin | Drug Info | Phase 3 | Solid tumour/cancer | [2], [3] | |
| 2 | PMX-53 | Drug Info | Phase 2 | Atopic dermatitis | [4], [5] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 58 Inhibitor drugs | + | ||||
| 1 | Curcumin | Drug Info | [1] | |||
| 2 | PMX-53 | Drug Info | [6] | |||
| 3 | (-)-clusin | Drug Info | [7] | |||
| 4 | (-)-cubebin | Drug Info | [7] | |||
| 5 | (-)-cubebinin | Drug Info | [7] | |||
| 6 | (-)-cubebininolide | Drug Info | [7] | |||
| 7 | (-)-dihydroclusin | Drug Info | [7] | |||
| 8 | (-)-thujaplicatintrimethyl ether | Drug Info | [7] | |||
| 9 | (-)-yatein | Drug Info | [7] | |||
| 10 | (8R,8'R)-4-hydroxycubebinone | Drug Info | [7] | |||
| 11 | (8R,8'R,9'S)-5-methoxyclusin | Drug Info | [7] | |||
| 12 | 1,1':4',1''-terphenyl-3,3''-diol | Drug Info | [8] | |||
| 13 | 1,2-Diacetoxylycorine | Drug Info | [9] | |||
| 14 | 1-(3,4-DICHLOROPHENYL)-6-(METHOXYMETHYL)-3-AZABICYCLO[4.1.0]HEPTANE (ENANTIOMERIC MIX) | Drug Info | [10] | |||
| 15 | 1-(4-Butoxy-phenyl)-1H-imidazole | Drug Info | [11] | |||
| 16 | 1-(METHOXYMETHYL)-6-(NAPHTHALEN-2-YL)-3-AZABICYCLO[4.1.0]HEPTANE (ENANTIOMERIC MIX) | Drug Info | [10] | |||
| 17 | 1-Acetoxy-2-tert-butyldimethylsilyl-oxylycorine | Drug Info | [9] | |||
| 18 | 1-Acetoxylycorine | Drug Info | [9] | |||
| 19 | 1H-1,2,3-benzotriazol-1-amine | Drug Info | [12] | |||
| 20 | 2,5-bis(4-hydroxybenzylidene)cyclopentanone | Drug Info | [1] | |||
| 21 | 2-(2-(4-tert-Butylphenylthio)ethyl)-1H-imidazole | Drug Info | [13] | |||
| 22 | 2-(4-Imidazol-1-yl-phenoxymethyl)-pyridine | Drug Info | [11] | |||
| 23 | 2-Fluoro-4-[5-(3-hydroxyphenyl)-2-thienyl]phenol | Drug Info | [14] | |||
| 24 | 2-tert-Butyldimethylsilyloxylycorine | Drug Info | [9] | |||
| 25 | 3,3-(1,3-Thiazole-2,5-diyl)diphenol | Drug Info | [8] | |||
| 26 | 3-(3-methylthiophen-2-yl)pyridine | Drug Info | [15] | |||
| 27 | 3-(5-((methylthio)methyl)furan-2-yl)pyridine | Drug Info | [15] | |||
| 28 | 3-(5-[1,3]dithiolan-2-yl-furan-2-yl)pyridine | Drug Info | [15] | |||
| 29 | 3-(pyridin-3-yl)prop-2-yn-1-amine | Drug Info | [15] | |||
| 30 | 3-methoxy-5-(6-methoxynaphthalen-2-yl)pyridine | Drug Info | [16] | |||
| 31 | 3-[3-(4-Imidazol-1-yl-phenoxy)-propyl]-pyridine | Drug Info | [11] | |||
| 32 | 3-[3-(4-Methoxybenzyl)naphthalen-2-yl]pyridine | Drug Info | [17] | |||
| 33 | 3-[5-(4-Hydroxyphenyl)-3-thienyl]phenol | Drug Info | [8] | |||
| 34 | 4-(6-Methoxynaphthalen-2-yl)isoquinoline | Drug Info | [16] | |||
| 35 | 4-[2-(4-Imidazol-1-yl-phenoxy)-ethyl]-morpholine | Drug Info | [11] | |||
| 36 | 4-[3-(4-Imidazol-1-yl-phenoxy)-propyl]-pyridine | Drug Info | [11] | |||
| 37 | 4-[5-(3-Hydroxyphenyl)-3-thienyl]-2-methylphenol | Drug Info | [14] | |||
| 38 | 5-(2-phenethylpiperazin-1-yl)-1H-indazole | Drug Info | [18] | |||
| 39 | 5-Pyridin-3-yl-thiophene-2-carbaldehyde oxime | Drug Info | [19] | |||
| 40 | 6-(3,4-DICHLOROPHENYL)-1-[1-(METHYLOXY)-3-BUTEN-1-YL]-3-AZABICYCLO[4.1.0]HEPTANE (DIASTEREOMERIC MIX) | Drug Info | [10] | |||
| 41 | 6-(3-Hydroxy-phenyl)-naphthalen-2-ol | Drug Info | [20] | |||
| 42 | 6-Pyridin-3-yl-3,4-dihydronaphthalen-2(1H)-one | Drug Info | [21] | |||
| 43 | Alpha-Hydroxy-Midazolam | Drug Info | [22] | |||
| 44 | Alpha-methylcubebin | Drug Info | [7] | |||
| 45 | BERGAPTOL | Drug Info | [23] | |||
| 46 | BMS-536924 | Drug Info | [24] | |||
| 47 | DIETHOXYFLOURESCEIN | Drug Info | [25] | |||
| 48 | DIHYDROCUBEBIN | Drug Info | [7] | |||
| 49 | ETHOXYCLUSIN | Drug Info | [7] | |||
| 50 | Geranylcoumarin | Drug Info | [23] | |||
| 51 | Go-Y026 | Drug Info | [1] | |||
| 52 | GSK-8062 | Drug Info | [26] | |||
| 53 | MEDIORESINOL | Drug Info | [7] | |||
| 54 | Methyl-(5-pyridin-3-yl-thiophen-2-yl)-amine | Drug Info | [19] | |||
| 55 | ML-3163 | Drug Info | [27] | |||
| 56 | ML-3375 | Drug Info | [27] | |||
| 57 | PALMATINE | Drug Info | [28] | |||
| 58 | TILIROSIDE | Drug Info | [29] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Bromocriptine | Ligand Info | |||||
| Structure Description | Crystal structure of the cytochrome P4503A4-bromoergocryptine complex | PDB:3UA1 | ||||
| Method | X-ray diffraction | Resolution | 2.15 Å | Mutation | No | [30] |
| PDB Sequence |
HSHGLFKKLG
37 IPGPTPLPFL47 GNILSYHKGF57 CMFDMECHKK67 YGKVWGFYDG77 QQPVLAITDP 87 DMIKTVLVKE97 CYSVFTNRRP107 FGPVGFMKSA117 ISIAEDEEWK127 RLRSLLSPTF 137 TSGKLKEMVP147 IIAQYGDVLV157 RNLRREAETG167 KPVTLKDVFG177 AYSMDVITST 187 SFGVNIDSLN197 NPQDPFVENT207 KKLLRFDFLD217 PFFLSITVFP227 FLIPILEVLN 237 ICVFPREVTN247 FLRKSVKRMK257 EEDTQVDFLQ273 LMIDSQHKAL290 SDLELVAQSI 300 IFIFAGYETT310 SSVLSFIMYE320 LATHPDVQQK330 LQEEIDAVLP340 NKAPPTYDTV 350 LQMEYLDMVV360 NETLRLFPIA370 MRLERVCKKD380 VEINGMFIPK390 GVVVMIPSYA 400 LHRDPKYWTE410 PEKFLPERFS420 KKNKDNIDPY430 IYTPFGSGPR440 NCIGMRFALM 450 NMKLALIRVL460 QNFSFKPCKE470 TQIPLKLSLG480 GLLQPEKPVV490 LKVESR |
|||||
|
|
TYR53
3.707
PHE57
3.569
ASP76
3.547
ARG105
3.267
ARG106
3.678
PRO107
3.820
PHE108
3.750
SER119
3.317
ILE120
3.431
ARG212
3.732
PHE213
4.413
PHE215
3.554
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Progesterone | Ligand Info | |||||
| Structure Description | Crystal structure of cytochrome P450 3A4 bound to progesterone | PDB:5A1R | ||||
| Method | X-ray diffraction | Resolution | 2.45 Å | Mutation | No | [31] |
| PDB Sequence |
GTHSHGLFKK
35 LGIPGPTPLP45 FLGNILSYHK55 GFCMFDMECH65 KKYGKVWGFY75 DGQQPVLAIT 85 DPDMIKTVLV95 KECYSVFTNR105 RPFGPVGFMK115 SAISIAEDEE125 WKRLRSLLSP 135 TFTSGKLKEM145 VPIIAQYGDV155 LVRNLRREAE165 TGKPVTLKDV175 FGAYSMDVIT 185 STSFGVNIDS195 LNNPQDPFVE205 NTKKLLRFDF215 LDPFFLSITV225 FPFLIPILEV 235 LNICVFPREV245 TNFLRKSVKR255 MKESRLERVD270 FLQLMIDSQN280 SHKALSDLEL 295 VAQSIIFIFA305 GYETTSSVLS315 FIMYELATHP325 DVQQKLQEEI335 DAVLPNKAPP 345 TYDTVLQMEY355 LDMVVNETLR365 LFPIAMRLER375 VCKKDVEING385 MFIPKGVVVM 395 IPSYALHRDP405 KYWTEPEKFL415 PERFSKKNKD425 NIDPYIYTPF435 GSGPRNCIGM 445 RFALMNMKLA455 LIRVLQNFSF465 KPCKETQIPL475 KLSLGGLLQP485 EKPVVLKVES 495 RD
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Steroid hormone biosynthesis | hsa00140 | Affiliated Target |
|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
| Linoleic acid metabolism | hsa00591 | Affiliated Target |
|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
| Retinol metabolism | hsa00830 | Affiliated Target |
|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Metabolism of xenobiotics by cytochrome P450 | hsa00980 | Affiliated Target |
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Drug metabolism - cytochrome P450 | hsa00982 | Affiliated Target |
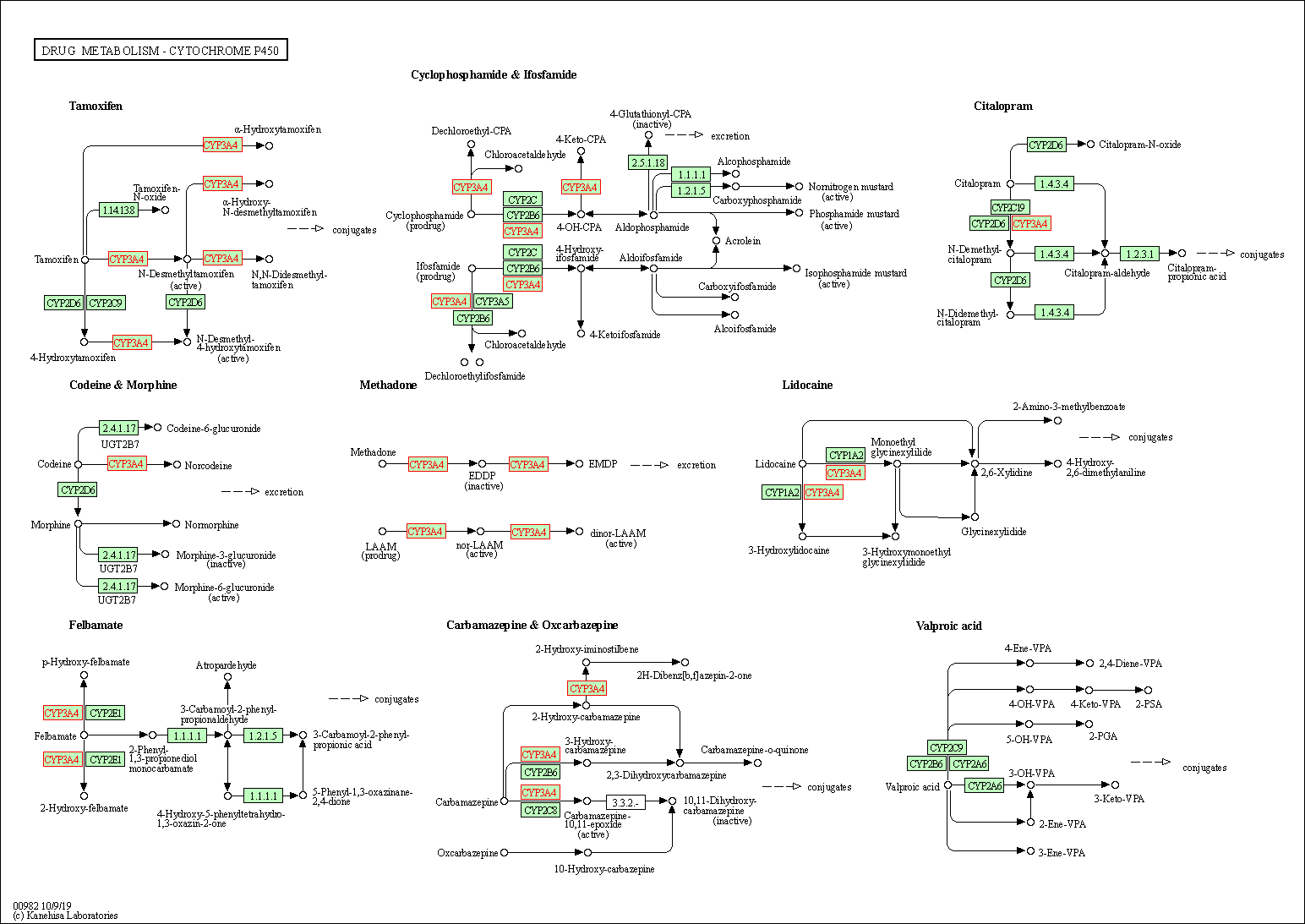
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Drug metabolism - other enzymes | hsa00983 | Affiliated Target |
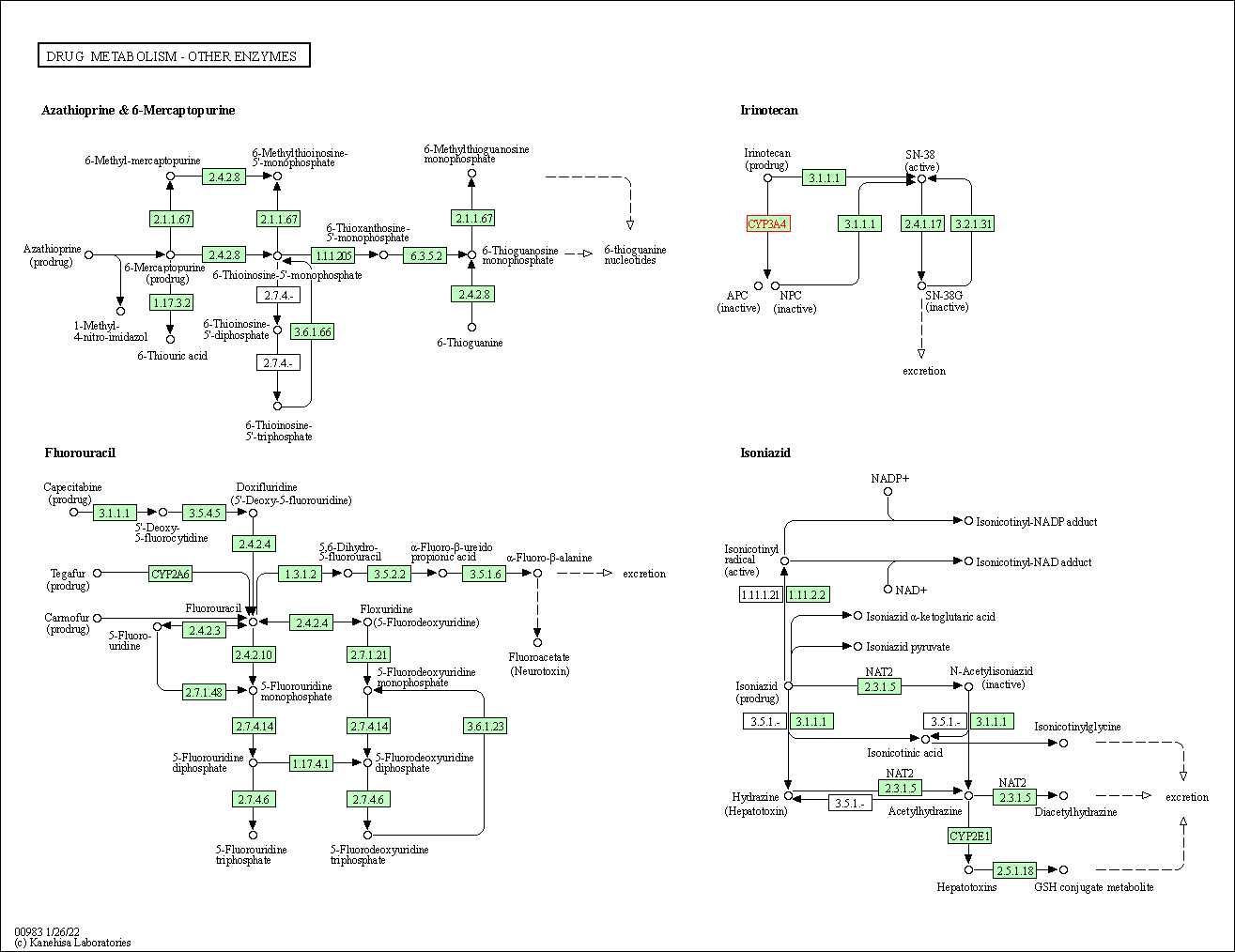
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Bile secretion | hsa04976 | Affiliated Target |
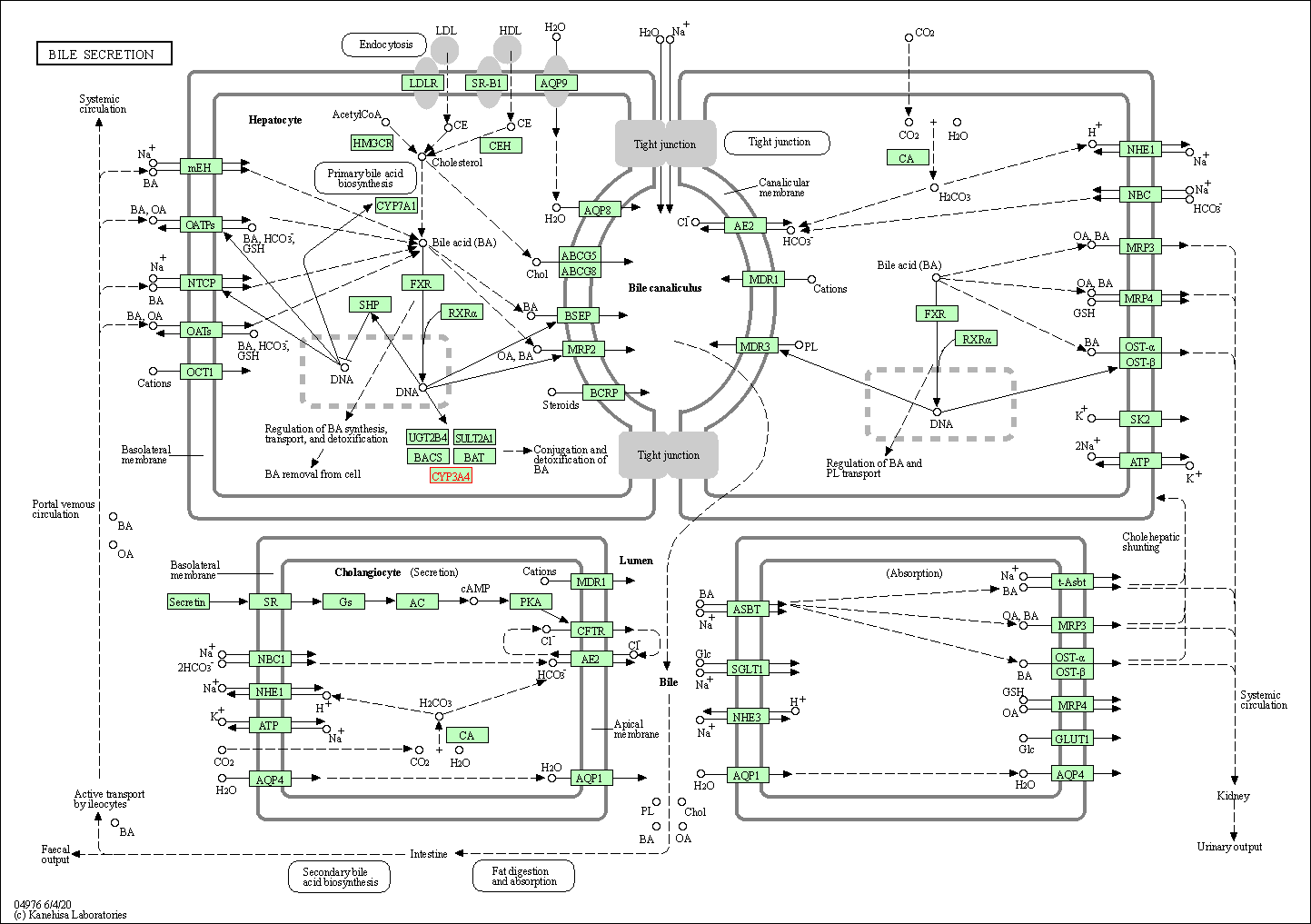
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 20 | Degree centrality | 2.15E-03 | Betweenness centrality | 3.06E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 1.93E-01 | Radiality | 1.33E+01 | Clustering coefficient | 4.74E-02 |
| Neighborhood connectivity | 4.15E+00 | Topological coefficient | 9.00E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| Target QSAR Model | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Structure-activity relationships for the inhibition of recombinant human cytochromes P450 by curcumin analogues. Eur J Med Chem. 2008 Aug;43(8):1621-31. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7000). | |||||
| REF 3 | Nanocurcumin: a promising therapeutic advancement over native curcumin. Crit Rev Ther Drug Carrier Syst. 2013;30(4):331-68. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 579). | |||||
| REF 5 | PMX-53 as a dual CD88 antagonist and an agonist for Mas-related gene 2 (MrgX2) in human mast cells. Mol Pharmacol. 2011 Jun;79(6):1005-13. | |||||
| REF 6 | Peptidomimetic C5a receptor antagonists with hydrophobic substitutions at the C-terminus: increased receptor specificity and in vivo activity. Bioorg Med Chem Lett. 2006 Oct 1;16(19):5088-92. | |||||
| REF 7 | Potent CYP3A4 inhibitory constituents of Piper cubeba. J Nat Prod. 2005 Jan;68(1):64-8. | |||||
| REF 8 | Design, synthesis, biological evaluation and pharmacokinetics of bis(hydroxyphenyl) substituted azoles, thiophenes, benzenes, and aza-benzenes as p... J Med Chem. 2008 Nov 13;51(21):6725-39. | |||||
| REF 9 | Selective cytochrome P450 3A4 inhibitory activity of Amaryllidaceae alkaloids. Bioorg Med Chem Lett. 2009 Jun 15;19(12):3233-7. | |||||
| REF 10 | 6-(3,4-dichlorophenyl)-1-[(methyloxy)methyl]-3-azabicyclo[4.1.0]heptane: a new potent and selective triple reuptake inhibitor. J Med Chem. 2010 Jul 8;53(13):4989-5001. | |||||
| REF 11 | Imidazole derivatives as new potent and selective 20-HETE synthase inhibitors. Bioorg Med Chem Lett. 2004 Jan 19;14(2):333-6. | |||||
| REF 12 | Discovery of a N'-hydroxyphenylformamidine derivative HET0016 as a potent and selective 20-HETE synthase inhibitor. Bioorg Med Chem Lett. 2001 Dec 3;11(23):2993-5. | |||||
| REF 13 | Role of hydrophobic substituents on the terminal nitrogen of histamine in receptor binding and agonist activity: development of an orally active hi... J Med Chem. 2010 May 13;53(9):3840-4. | |||||
| REF 14 | New insights into the SAR and binding modes of bis(hydroxyphenyl)thiophenes and -benzenes: influence of additional substituents on 17beta-hydroxyst... J Med Chem. 2009 Nov 12;52(21):6724-43. | |||||
| REF 15 | Synthetic inhibitors of cytochrome P-450 2A6: inhibitory activity, difference spectra, mechanism of inhibition, and protein cocrystallization. J Med Chem. 2006 Nov 30;49(24):6987-7001. | |||||
| REF 16 | Overcoming undesirable CYP1A2 inhibition of pyridylnaphthalene-type aldosterone synthase inhibitors: influence of heteroaryl derivatization on pote... J Med Chem. 2008 Aug 28;51(16):5064-74. | |||||
| REF 17 | Novel aldosterone synthase inhibitors with extended carbocyclic skeleton by a combined ligand-based and structure-based drug design approach. J Med Chem. 2008 Oct 9;51(19):6138-49. | |||||
| REF 18 | 2-Substituted N-aryl piperazines as novel triple reuptake inhibitors for the treatment of depression, Bioorg. Med. Chem. Lett. 20(13):3941-3945 (2010). | |||||
| REF 19 | 5-substituted, 6-substituted, and unsubstituted 3-heteroaromatic pyridine analogues of nicotine as selective inhibitors of cytochrome P-450 2A6. J Med Chem. 2005 Jan 13;48(1):224-39. | |||||
| REF 20 | Design, synthesis, and biological evaluation of (hydroxyphenyl)naphthalene and -quinoline derivatives: potent and selective nonsteroidal inhibitors... J Med Chem. 2008 Apr 10;51(7):2158-69. | |||||
| REF 21 | In vivo active aldosterone synthase inhibitors with improved selectivity: lead optimization providing a series of pyridine substituted 3,4-dihydro-... J Med Chem. 2008 Dec 25;51(24):8077-87. | |||||
| REF 22 | Design and synthesis of a new fluorescent probe for cytochrome P450 3A4 (CYP 3A4). Bioorg Med Chem Lett. 2003 Nov 3;13(21):3643-5. | |||||
| REF 23 | Radical scavenging and cytochrome P450 3A4 inhibitory activity of bergaptol and geranylcoumarin from grapefruit. Bioorg Med Chem. 2007 Jun 1;15(11):3684-91. | |||||
| REF 24 | Discovery and evaluation of 4-(2-(4-chloro-1H-pyrazol-1-yl)ethylamino)-3-(6-(1-(3-fluoropropyl)piperidin-4-yl)-4-methyl-1H-benzo[d]imidazol-2-yl)py... J Med Chem. 2008 Oct 9;51(19):5897-900. | |||||
| REF 25 | Exploration of the amine terminus in a novel series of 1,2,4-triazolo-3-yl-azabicyclo[3.1.0]hexanes as selective dopamine D3 receptor antagonists. J Med Chem. 2010 Oct 14;53(19):7129-39. | |||||
| REF 26 | Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064. Bioorg Med Chem Lett. 2008 Aug 1;18(15):4339-43. | |||||
| REF 27 | Novel substituted pyridinyl imidazoles as potent anticytokine agents with low activity against hepatic cytochrome P450 enzymes. J Med Chem. 2003 Jul 17;46(15):3230-44. | |||||
| REF 28 | Cytochrome P3A4 inhibitors and other constituents of Fibraurea tinctoria. J Nat Prod. 2007 Dec;70(12):1930-3. | |||||
| REF 29 | Isolation of cytochrome P450 inhibitors from strawberry fruit, Fragaria ananassa. J Nat Prod. 2004 Nov;67(11):1839-41. | |||||
| REF 30 | Structural and mechanistic insights into the interaction of cytochrome P4503A4 with bromoergocryptine, a type I ligand. J Biol Chem. 2012 Jan 27;287(5):3510-7. | |||||
| REF 31 | Anion-Dependent Stimulation of CYP3A4 Monooxygenase. Biochemistry. 2015 Jul 7;54(26):4083-96. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

