Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T43115
(Former ID: TTDR00611)
|
|||||
| Target Name |
Leukocyte common antigen (PTPRC)
|
|||||
| Synonyms |
T200; Receptor-type tyrosine-protein phosphatase C; L-CA; CD45 antigen; CD45
Click to Show/Hide
|
|||||
| Gene Name |
PTPRC
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Acute myeloid leukaemia [ICD-11: 2A60] | |||||
| 2 | Transplanted organ/tissue [ICD-11: QB63] | |||||
| Function |
Acts as a positive regulator of T-cell coactivation upon binding to DPP4. The first PTPase domain has enzymatic activity, while the second one seems to affect the substrate specificity of the first one. Upon T-cell activation, recruits and dephosphorylates SKAP1 and FYN. Dephosphorylates LYN, and thereby modulates LYN activity. Protein tyrosine-protein phosphatase required for T-cell activation through the antigen receptor.
Click to Show/Hide
|
|||||
| BioChemical Class |
Phosphoric monoester hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.1.3.48
|
|||||
| Sequence |
MTMYLWLKLLAFGFAFLDTEVFVTGQSPTPSPTGLTTAKMPSVPLSSDPLPTHTTAFSPA
STFERENDFSETTTSLSPDNTSTQVSPDSLDNASAFNTTGVSSVQTPHLPTHADSQTPSA GTDTQTFSGSAANAKLNPTPGSNAISDVPGERSTASTFPTDPVSPLTTTLSLAHHSSAAL PARTSNTTITANTSDAYLNASETTTLSPSGSAVISTTTIATTPSKPTCDEKYANITVDYL YNKETKLFTAKLNVNENVECGNNTCTNNEVHNLTECKNASVSISHNSCTAPDKTLILDVP PGVEKFQLHDCTQVEKADTTICLKWKNIETFTCDTQNITYRFQCGNMIFDNKEIKLENLE PEHEYKCDSEILYNNHKFTNASKIIKTDFGSPGEPQIIFCRSEAAHQGVITWNPPQRSFH NFTLCYIKETEKDCLNLDKNLIKYDLQNLKPYTKYVLSLHAYIIAKVQRNGSAAMCHFTT KSAPPSQVWNMTVSMTSDNSMHVKCRPPRDRNGPHERYHLEVEAGNTLVRNESHKNCDFR VKDLQYSTDYTFKAYFHNGDYPGEPFILHHSTSYNSKALIAFLAFLIIVTSIALLVVLYK IYDLHKKRSCNLDEQQELVERDDEKQLMNVEPIHADILLETYKRKIADEGRLFLAEFQSI PRVFSKFPIKEARKPFNQNKNRYVDILPYDYNRVELSEINGDAGSNYINASYIDGFKEPR KYIAAQGPRDETVDDFWRMIWEQKATVIVMVTRCEEGNRNKCAEYWPSMEEGTRAFGDVV VKINQHKRCPDYIIQKLNIVNKKEKATGREVTHIQFTSWPDHGVPEDPHLLLKLRRRVNA FSNFFSGPIVVHCSAGVGRTGTYIGIDAMLEGLEAENKVDVYGYVVKLRRQRCLMVQVEA QYILIHQALVEYNQFGETEVNLSELHPYLHNMKKRDPPSEPSPLEAEFQRLPSYRSWRTQ HIGNQEENKSKNRNSNVIPYDYNRVPLKHELEMSKESEHDSDESSDDDSDSEEPSKYINA SFIMSYWKPEVMIAAQGPLKETIGDFWQMIFQRKVKVIVMLTELKHGDQEICAQYWGEGK QTYGDIEVDLKDTDKSSTYTLRVFELRHSKRKDSRTVYQYQYTNWSVEQLPAEPKELISM IQVVKQKLPQKNSSEGNKHHKSTPLLIHCRDGSQQTGIFCALLNLLESAETEEVVDIFQV VKALRKARPGMVSTFEQYQFLYDVIASTYPAQNGQVKKNNHQEDKIEFDNEVDKVKQDAN CVNPLGAPEKLPEAKEQAEGSEPTSGTEGPEHSVNGPASPALNQGS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | Iomab-B | Drug Info | Phase 3 | Bone marrow transplantation | [3] | |
| 2 | [131I]-BC8 | Drug Info | Phase 2 | leukaemia | [4] | |
| 3 | Anti-CD45 mabs | Drug Info | Phase 1 | Lymphoma | [6] | |
| 4 | LM-CD45 | Drug Info | Phase 1 | Transplant rejection | [7] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | Asp-BrPmp-Leu | Drug Info | [12] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cell adhesion molecules | hsa04514 | Affiliated Target |
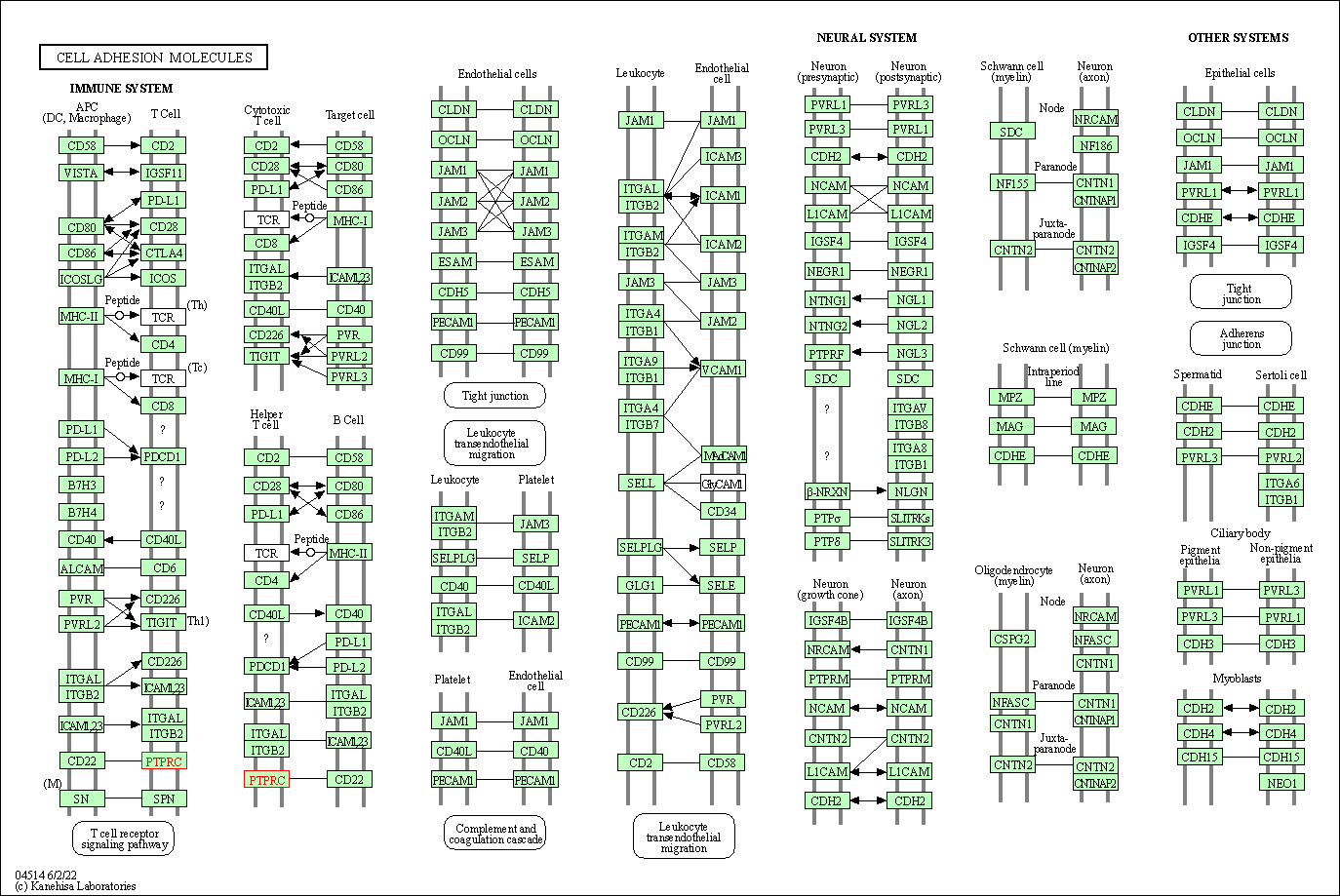
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |
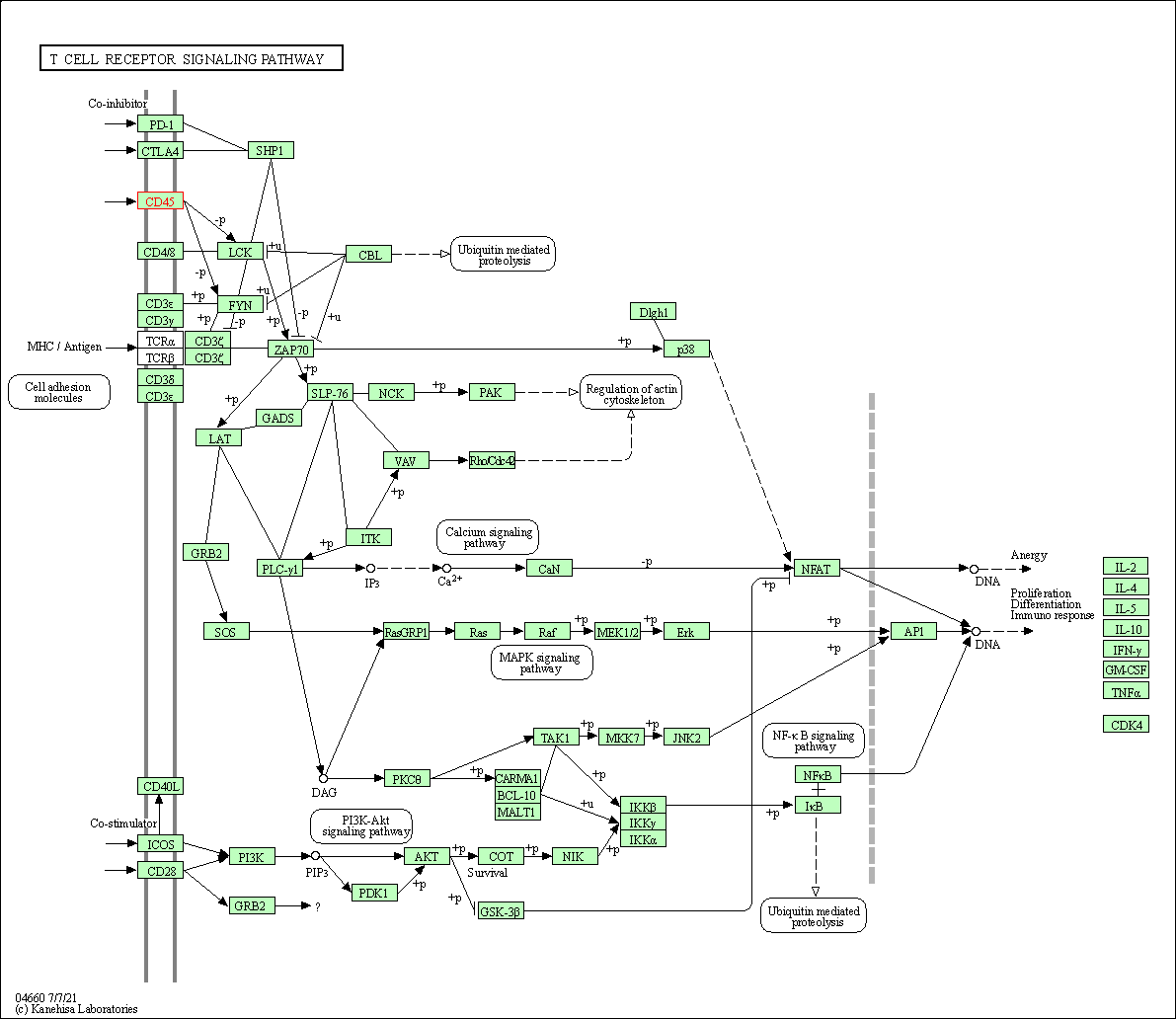
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc gamma R-mediated phagocytosis | hsa04666 | Affiliated Target |
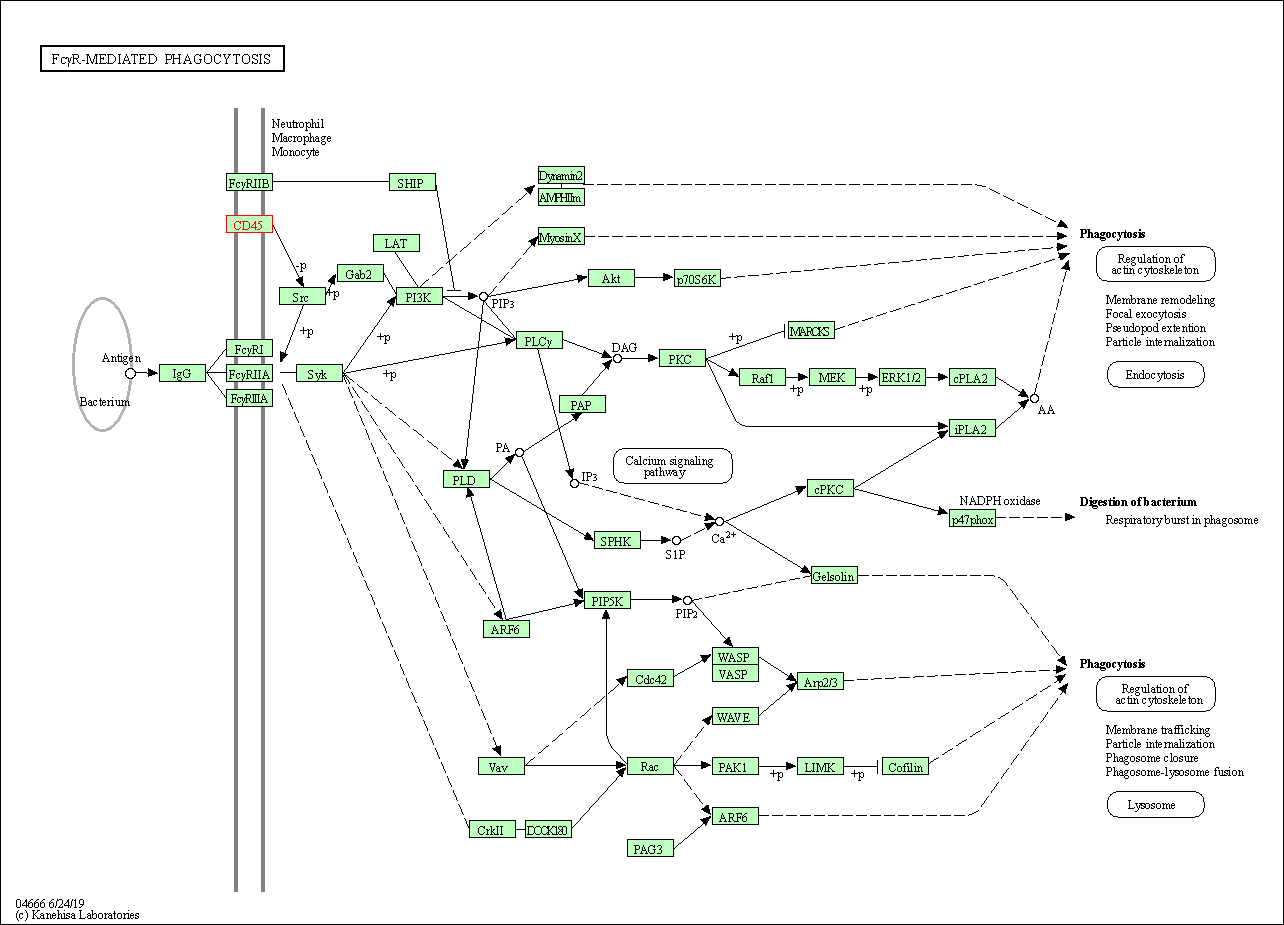
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 23 | Degree centrality | 2.47E-03 | Betweenness centrality | 8.17E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.27E-01 | Radiality | 1.40E+01 | Clustering coefficient | 1.74E-01 |
| Neighborhood connectivity | 2.46E+01 | Topological coefficient | 8.87E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Cell adhesion molecules (CAMs) | |||||
| 2 | T cell receptor signaling pathway | |||||
| 3 | Fc gamma R-mediated phagocytosis | |||||
| 4 | Primary immunodeficiency | |||||
| NetPath Pathway | [+] 2 NetPath Pathways | + | ||||
| 1 | TCR Signaling Pathway | |||||
| 2 | IL2 Signaling Pathway | |||||
| Panther Pathway | [+] 3 Panther Pathways | + | ||||
| 1 | B cell activation | |||||
| 2 | JAK/STAT signaling pathway | |||||
| 3 | T cell activation | |||||
| PID Pathway | [+] 6 PID Pathways | + | ||||
| 1 | BCR signaling pathway | |||||
| 2 | TCR signaling in naï | |||||
| 3 | ||||||
| 4 | TCR signaling in naï | |||||
| 5 | ||||||
| 6 | CXCR4-mediated signaling events | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Phosphorylation of CD3 and TCR zeta chains | |||||
| 2 | Other semaphorin interactions | |||||
| WikiPathways | [+] 5 WikiPathways | + | ||||
| 1 | Interferon type I signaling pathways | |||||
| 2 | EPO Receptor Signaling | |||||
| 3 | B Cell Receptor Signaling Pathway | |||||
| 4 | TCR signaling | |||||
| 5 | Semaphorin interactions | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of Actinium Pharmaceuticals. | |||||
| REF 2 | ClinicalTrials.gov (NCT02665065) Study of Iomab-B vs. Conventional Care in Older Subjects With Active, Relapsed or Refractory Acute Myeloid Leukemia (SIERRA). U.S. National Institutes of Health. | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | Development of a marrow transplant regimen for acute leukemia using targeted hematopoietic irradiation delivered by 131I-labeled anti-CD45 antibody, combined with cyclophosphamide and total body irradiation. Blood. 1995 Feb 15;85(4):1122-31. | |||||
| REF 5 | ClinicalTrials.gov (NCT04512716) Iomab-ACT: A Pilot Study of 131-I Apamistamab Followed by CD19-Targeted CAR T-Cell Therapy for Patients With Relapsed or Refractory B-Cell Acute Lymphoblastic Leukemia or Diffuse Large B-Cell Lymphoma. U.S. National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT00078546) EBV-Specific CTLs Following CD45 Antibody to Patients With Epstein-Barr Virus (EBV) + Nasopharyngeal Carcinoma (NPC). U.S. National Institutes of Health. | |||||
| REF 7 | ClinicalTrials.gov (NCT01678443) Monoclonal Antibody Therapy Before Stem Cell Transplant in Treating Patients With Relapsed or Refractory Lymphoid Malignancies. U.S. National Institutes of Health. | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800027164) | |||||
| REF 9 | Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012 Jan;40(Database issue):D1128-36. | |||||
| REF 10 | CD45 monoclonal antibody-mediated cytolysis of human NK and T lymphoma cells. Haematologica. 2006 Jul;91(7):886-94. | |||||
| REF 11 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002932) | |||||
| REF 12 | A protected l-bromophosphonomethylphenylalanine amino acid derivative (BrPmp) for synthesis of irreversible protein tyrosine phosphatase inhibitors. Bioorg Med Chem. 2010 Dec 15;18(24):8679-86. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

