Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T44861
(Former ID: TTDNC00553)
|
|||||
| Target Name |
Protein kinase C delta (PRKCD)
|
|||||
| Synonyms |
nPKC-delta; Tyrosine-protein kinase PRKCD; SDK1; Protein kinase C delta type catalytic subunit; Protein kinase C delta type; PKC-delta
Click to Show/Hide
|
|||||
| Gene Name |
PRKCD
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Myocardial infarction [ICD-11: BA41-BA43] | |||||
| Function |
Negatively regulates B cell proliferation and also has an important function in self-antigen induced B cell tolerance induction. Upon DNA damage, activates the promoter of the death-promoting transcription factor BCLAF1/Btf to trigger BCLAF1-mediated p53/TP53 gene transcription and apoptosis. In response to oxidative stress, interact with and activate CHUK/IKKA in the nucleus, causing the phosphorylation of p53/TP53. In the case of ER stress or DNA damage-induced apoptosis, can form a complex with the tyrosine-protein kinase ABL1 which trigger apoptosis independently of p53/TP53. In cytosol can trigger apoptosis by activating MAPK11 or MAPK14, inhibiting AKT1 and decreasing the level of X-linked inhibitor of apoptosis protein (XIAP), whereas in nucleus induces apoptosis via the activation of MAPK8 or MAPK9. Upon ionizing radiation treatment, is required for the activation of the apoptosis regulators BAX and BAK, which trigger the mitochondrial cell death pathway. Can phosphorylate MCL1 and target it for degradation which is sufficient to trigger for BAX activation and apoptosis. Is required for the control of cell cycle progression both at G1/S and G2/M phases. Mediates phorbol 12-myristate 13-acetate (PMA)-induced inhibition of cell cycle progression at G1/S phase by up-regulating the CDK inhibitor CDKN1A/p21 and inhibiting the cyclin CCNA2 promoter activity. In response to UV irradiation can phosphorylate CDK1, which is important for the G2/M DNA damage checkpoint activation. Can protect glioma cells from the apoptosis induced by TNFSF10/TRAIL, probably by inducing increased phosphorylation and subsequent activation of AKT1. Is highly expressed in a number of cancer cells and promotes cell survival and resistance against chemotherapeutic drugs by inducing cyclin D1 (CCND1) and hyperphosphorylation of RB1, and via several pro-survival pathways, including NF-kappa-B, AKT1 and MAPK1/3 (ERK1/2). Can also act as tumor suppressor upon mitogenic stimulation with PMA or TPA. In N-formyl-methionyl-leucyl-phenylalanine (fMLP)-treated cells, is required for NCF1 (p47-phox) phosphorylation and activation of NADPH oxidase activity, and regulates TNF-elicited superoxide anion production in neutrophils, by direct phosphorylation and activation of NCF1 or indirectly through MAPK1/3 (ERK1/2) signaling pathways. May also play a role in the regulation of NADPH oxidase activity in eosinophil after stimulation with IL5, leukotriene B4 or PMA. In collagen-induced platelet aggregation, acts a negative regulator of filopodia formation and actin polymerization by interacting with and negatively regulating VASP phosphorylation. Downstream of PAR1, PAR4 and CD36/GP4 receptors, regulates differentially platelet dense granule secretion; acts as a positive regulator in PAR-mediated granule secretion, whereas it negatively regulates CD36/GP4-mediated granule release. Phosphorylates MUC1 in the C-terminal and regulates the interaction between MUC1 and beta-catenin. The catalytic subunit phosphorylates 14-3-3 proteins (YWHAB, YWHAZ and YWHAH) in a sphingosine-dependent fashion. Phosphorylates ELAVL1 in response to angiotensin-2 treatment. Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays contrasting roles in cell death and cell survival by functioning as a pro-apoptotic protein during DNA damage-induced apoptosis, but acting as an anti-apoptotic protein during cytokine receptor-initiated cell death, is involved in tumor suppression as well as survival of several cancers, is required for oxygen radical production by NADPH oxidase and acts as positive or negative regulator in platelet functional responses.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.13
|
|||||
| Sequence |
MAPFLRIAFNSYELGSLQAEDEANQPFCAVKMKEALSTERGKTLVQKKPTMYPEWKSTFD
AHIYEGRVIQIVLMRAAEEPVSEVTVGVSVLAERCKKNNGKAEFWLDLQPQAKVLMSVQY FLEDVDCKQSMRSEDEAKFPTMNRRGAIKQAKIHYIKNHEFIATFFGQPTFCSVCKDFVW GLNKQGYKCRQCNAAIHKKCIDKIIGRCTGTAANSRDTIFQKERFNIDMPHRFKVHNYMS PTFCDHCGSLLWGLVKQGLKCEDCGMNVHHKCREKVANLCGINQKLLAEALNQVTQRASR RSDSASSEPVGIYQGFEKKTGVAGEDMQDNSGTYGKIWEGSSKCNINNFIFHKVLGKGSF GKVLLGELKGRGEYFAIKALKKDVVLIDDDVECTMVEKRVLTLAAENPFLTHLICTFQTK DHLFFVMEFLNGGDLMYHIQDKGRFELYRATFYAAEIMCGLQFLHSKGIIYRDLKLDNVL LDRDGHIKIADFGMCKENIFGESRASTFCGTPDYIAPEILQGLKYTFSVDWWSFGVLLYE MLIGQSPFHGDDEDELFESIRVDTPHYPRWITKESKDILEKLFEREPTKRLGVTGNIKIH PFFKTINWTLLEKRRLEPPFRPKVKSPRDYSNFDQEFLNEKARLSYSDKNLIDSMDQSAF AGFSFVNPKFEHLLED Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A01328 ; BADD_A02014 ; BADD_A04282 ; BADD_A05675 | |||||
| HIT2.0 ID | T54G8J | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | KAI-9803 | Drug Info | Phase 1/2 | Acute myocardial infarction | [2], [3] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 21 Inhibitor drugs | + | ||||
| 1 | KAI-9803 | Drug Info | [1], [4], [5] | |||
| 2 | RO-320432 | Drug Info | [7] | |||
| 3 | 13-Acetylphorbol | Drug Info | [8] | |||
| 4 | 2,3,3-Triphenyl-acrylonitrile | Drug Info | [9] | |||
| 5 | 2-(4-Hydroxy-phenyl)-3,3-diphenyl-acrylonitrile | Drug Info | [9] | |||
| 6 | 3,3-Bis-(4-hydroxy-phenyl)-2-phenyl-acrylonitrile | Drug Info | [9] | |||
| 7 | 3,3-Bis-(4-methoxy-phenyl)-2-phenyl-acrylonitrile | Drug Info | [9] | |||
| 8 | 3-(4-Hydroxy-phenyl)-2,3-diphenyl-acrylonitrile | Drug Info | [9] | |||
| 9 | 4-cycloheptyliden(4-hydroxyphenyl)methylphenol | Drug Info | [9] | |||
| 10 | 4-[1-(4-hydroxyphenyl)-3-methyl-1-butenyl]phenol | Drug Info | [9] | |||
| 11 | INDOLACTUM | Drug Info | [10] | |||
| 12 | Ingenol-3-bezoate | Drug Info | [11] | |||
| 13 | LY-326449 | Drug Info | [12] | |||
| 14 | Phorbol 12,13-butyrate | Drug Info | [13] | |||
| 15 | PROSTRATIN | Drug Info | [14] | |||
| 16 | RO-316233 | Drug Info | [15] | |||
| 17 | Ro-32-0557 | Drug Info | [7] | |||
| 18 | THYMELEATOXIN | Drug Info | [11] | |||
| 19 | [2,2':5',2'']Terthiophen-4-yl-methanol | Drug Info | [16] | |||
| 20 | [2,2':5',2'']Terthiophene-4,5''-dicarbaldehyde | Drug Info | [16] | |||
| 21 | [2,2':5',2'']Terthiophene-4-carbaldehyde | Drug Info | [16] | |||
| Activator | [+] 1 Activator drugs | + | ||||
| 1 | AD 198 | Drug Info | [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Chemokine signaling pathway | hsa04062 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |
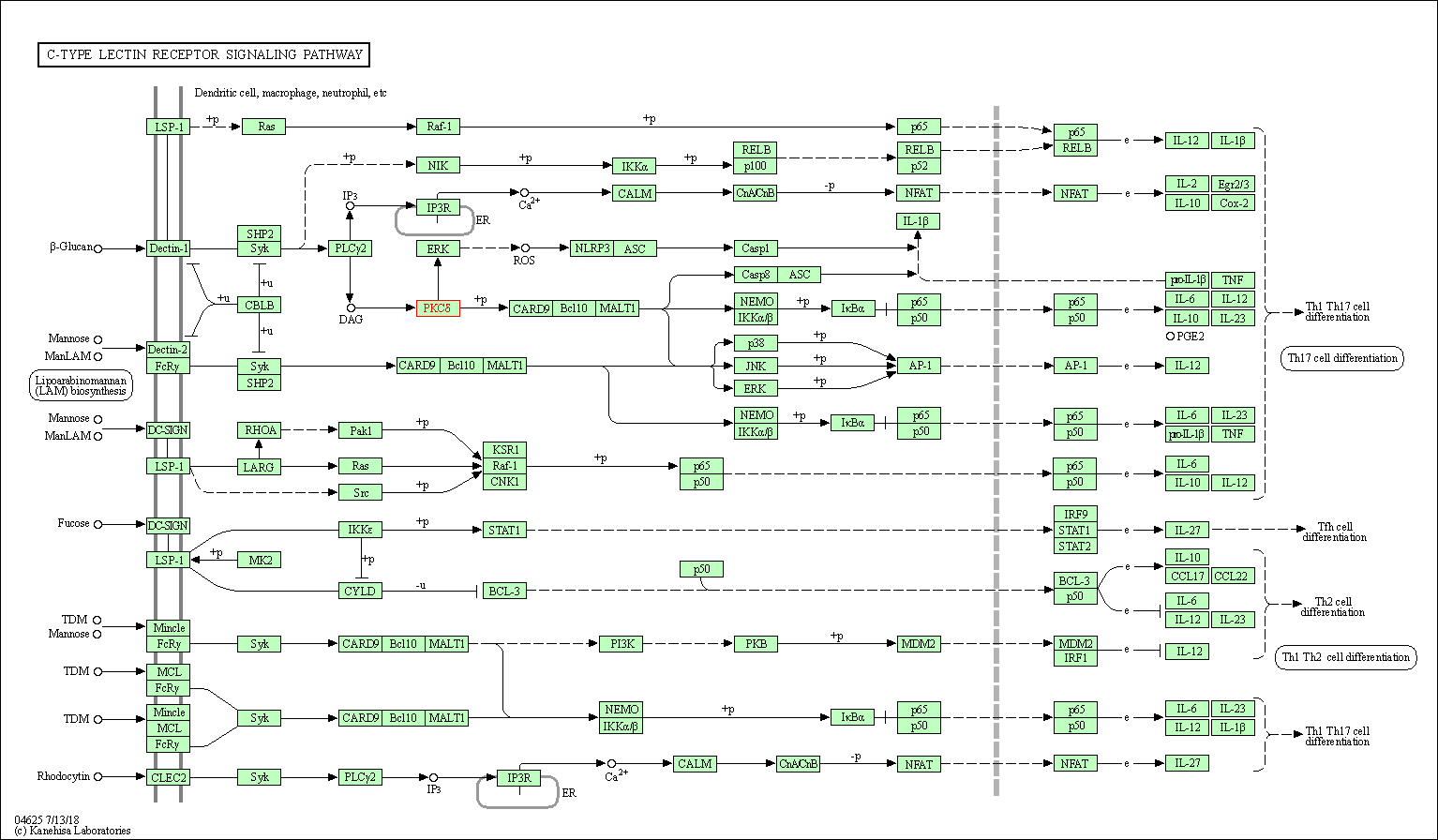
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc gamma R-mediated phagocytosis | hsa04666 | Affiliated Target |
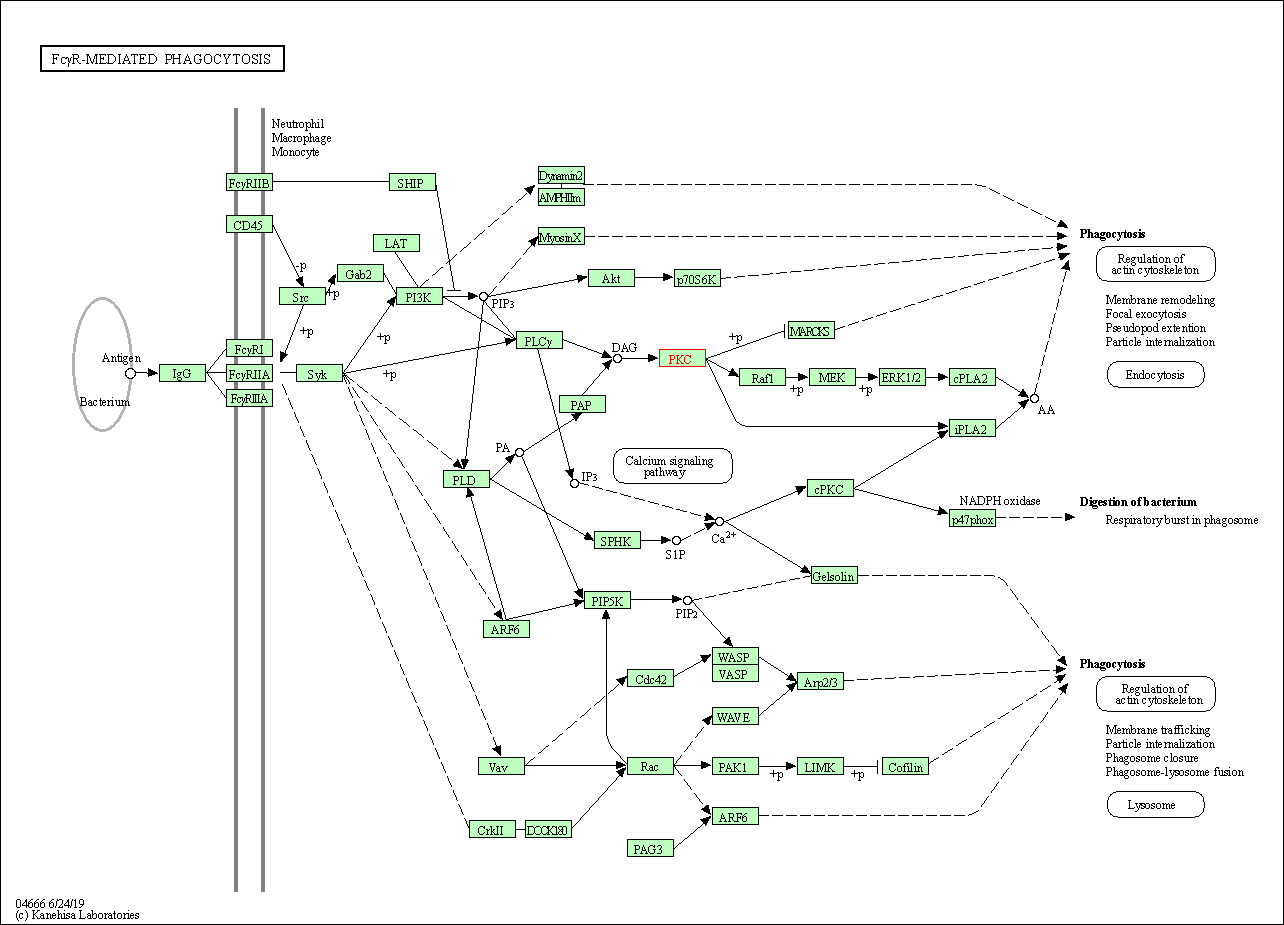
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |
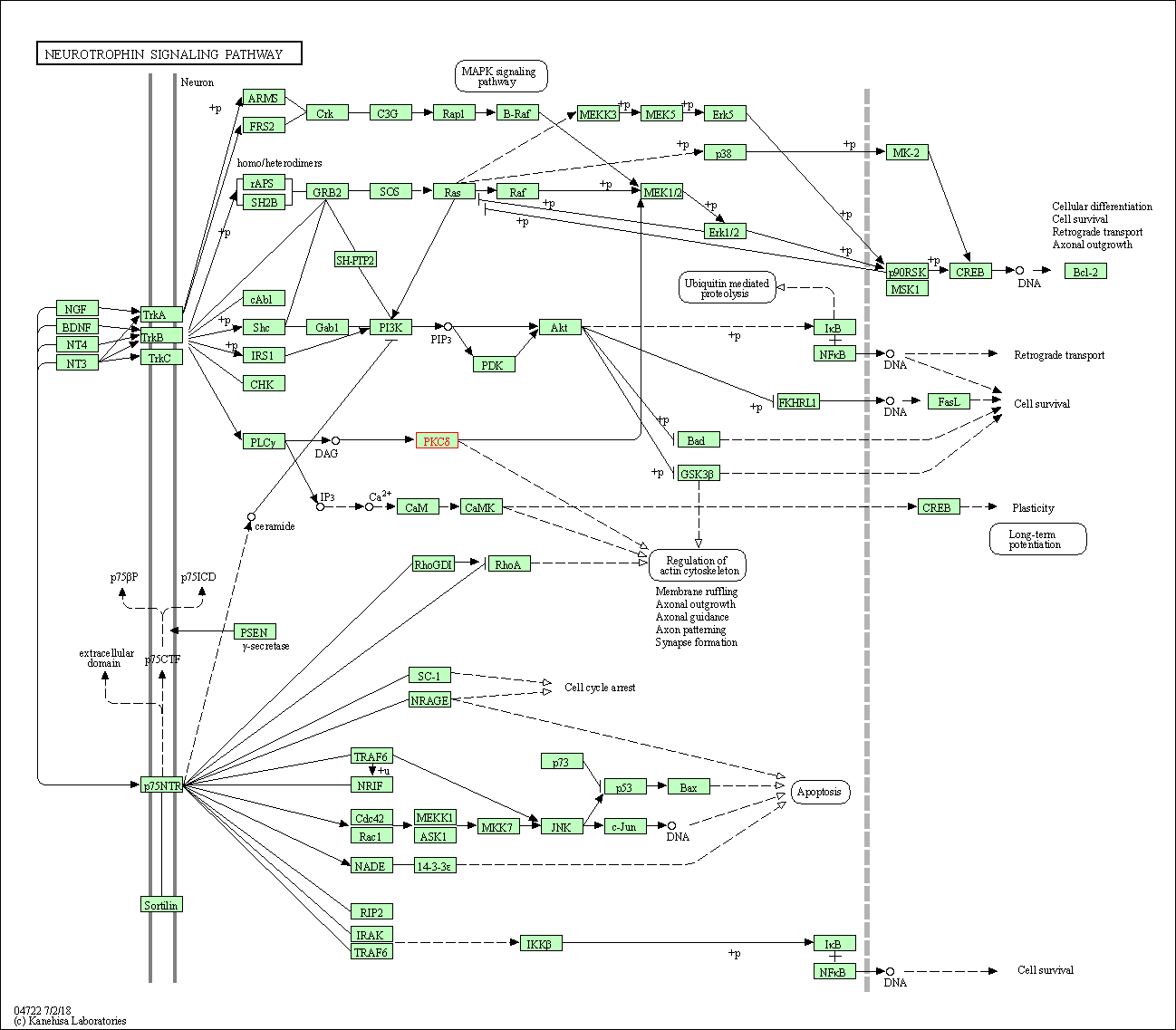
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |
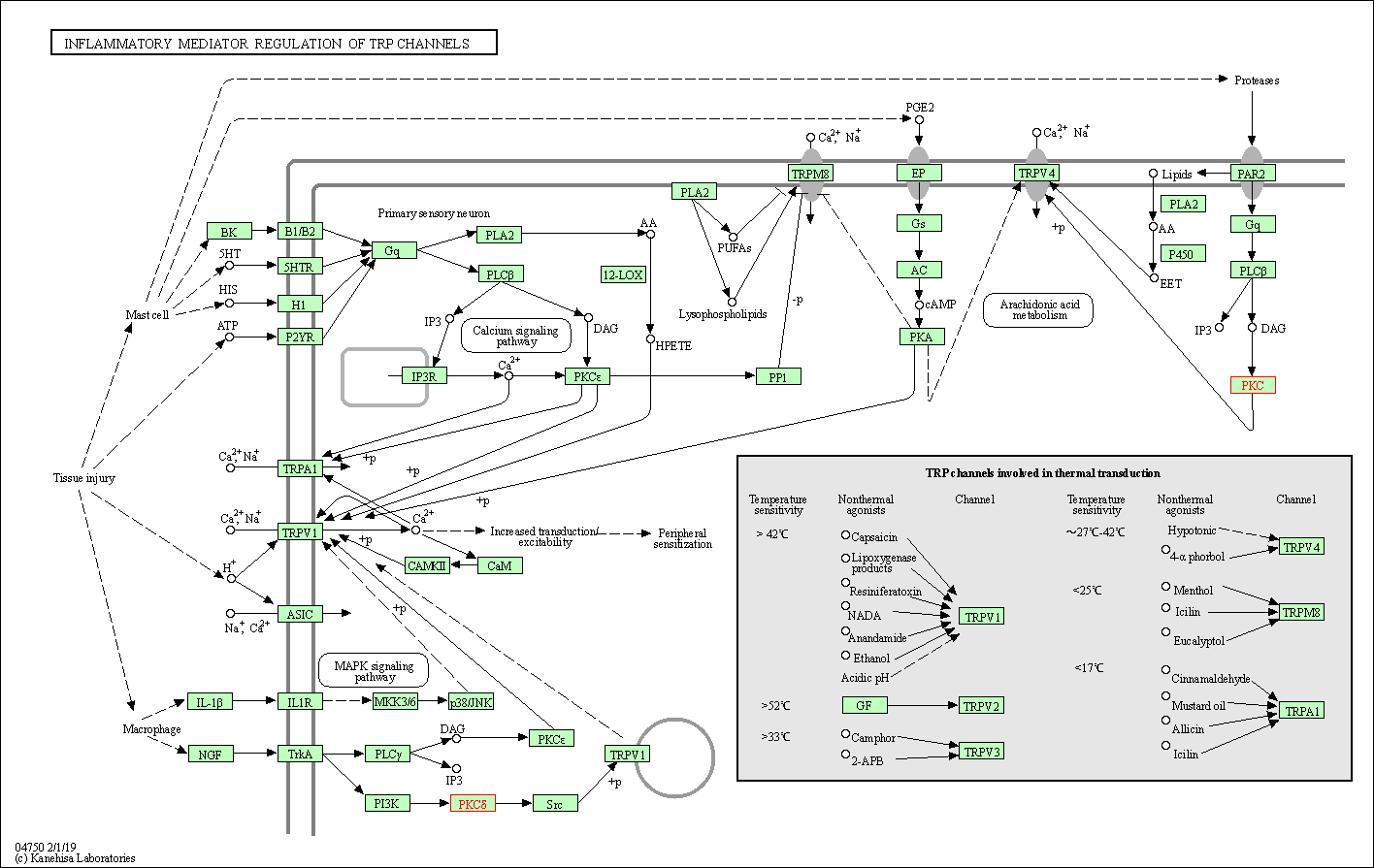
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 20 | Degree centrality | 2.15E-03 | Betweenness centrality | 1.00E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.62E-01 | Radiality | 1.45E+01 | Clustering coefficient | 1.95E-01 |
| Neighborhood connectivity | 7.01E+01 | Topological coefficient | 7.75E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | PKC delta and epsilon in drug targeting and therapeutics. Recent Pat DNA Gene Seq. 2009;3(2):96-101. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7840). | |||||
| REF 3 | Pharmacologic therapeutics for cardiac reperfusion injury. Expert Opin Emerg Drugs. 2007 Sep;12(3):367-88. | |||||
| REF 4 | Letter by Metzler et al regarding article, "Intracoronary KAI-9803 as an adjunct to primary coronary intervention for acute ST-segment elevation myocardial infarction". Circulation. 2008 Jul 22;118(4):e80. | |||||
| REF 5 | Intracoronary KAI-9803 as an adjunct to primary percutaneous coronary intervention for acute ST-segment elevation myocardial infarction. Circulation. 2008 Feb 19;117(7):886-96. | |||||
| REF 6 | Novel extranuclear-targeted anthracyclines override the antiapoptotic functions of Bcl-2 and target protein kinase C pathways to induce apoptosis. Mol Cancer Ther. 2002 May;1(7):469-81. | |||||
| REF 7 | Bisindolylmaleimide inhibitors of protein kinase C. Further conformational restriction of a tertiary amine side chain, Bioorg. Med. Chem. Lett. 4(11):1303-1308 (1994). | |||||
| REF 8 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 9 | Multivariate analysis by the minimum spanning tree method of the structural determinants of diphenylethylenes and triphenylacrylonitriles implicate... J Med Chem. 1992 Feb 7;35(3):573-83. | |||||
| REF 10 | Structural basis of RasGRP binding to high-affinity PKC ligands. J Med Chem. 2002 Feb 14;45(4):853-60. | |||||
| REF 11 | Structural basis of binding of high-affinity ligands to protein kinase C: prediction of the binding modes through a new molecular dynamics method a... J Med Chem. 2001 May 24;44(11):1690-701. | |||||
| REF 12 | (S)-13-[(dimethylamino)methyl]-10,11,14,15-tetrahydro-4,9:16, 21-dimetheno-1H, 13H-dibenzo[e,k]pyrrolo[3,4-h][1,4,13]oxadiazacyclohexadecene-1,3(2H... J Med Chem. 1996 Jul 5;39(14):2664-71. | |||||
| REF 13 | Conformationally constrained analogues of diacylglycerol (DAG). 28. DAG-dioxolanones reveal a new additional interaction site in the C1b domain of ... J Med Chem. 2007 Jul 26;50(15):3465-81. | |||||
| REF 14 | A nonpromoting phorbol from the samoan medicinal plant Homalanthus nutans inhibits cell killing by HIV-1. J Med Chem. 1992 May 29;35(11):1978-86. | |||||
| REF 15 | Inhibitors of protein kinase C. 1. 2,3-Bisarylmaleimides. J Med Chem. 1992 Jan;35(1):177-84. | |||||
| REF 16 | Novel protein kinase C inhibitors: synthesis and PKC inhibition of beta-substituted polythiophene derivatives. Bioorg Med Chem Lett. 1999 Aug 2;9(15):2279-82. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

