Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T49755
(Former ID: TTDNR00702)
|
|||||
| Target Name |
LDL receptor related protein-6 (LRP-6)
|
|||||
| Synonyms |
Low-density lipoprotein receptor-related protein 6
Click to Show/Hide
|
|||||
| Gene Name |
LRP6
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Cell-surface coreceptor of Wnt/beta-catenin signaling, which plays a pivotal role in bone formation. The Wnt-induced Fzd/LRP6 coreceptor complex recruits DVL1 polymers to the plasma membrane which, in turn, recruits the AXIN1/GSK3B-complex to the cell surface promoting the formation of signalsomes and inhibiting AXIN1/GSK3-mediated phosphorylation and destruction of beta-catenin. Required for posterior patterning of the epiblast during gastrulation. Component of the Wnt-Fzd-LRP5-LRP6 complex that triggers beta-catenin signaling through inducing aggregation of receptor-ligand complexes into ribosome-sized signalsomes.
Click to Show/Hide
|
|||||
| BioChemical Class |
Low density lipoprotein receptor
|
|||||
| UniProt ID | ||||||
| Sequence |
MGAVLRSLLACSFCVLLRAAPLLLYANRRDLRLVDATNGKENATIVVGGLEDAAAVDFVF
SHGLIYWSDVSEEAIKRTEFNKTESVQNVVVSGLLSPDGLACDWLGEKLYWTDSETNRIE VSNLDGSLRKVLFWQELDQPRAIALDPSSGFMYWTDWGEVPKIERAGMDGSSRFIIINSE IYWPNGLTLDYEEQKLYWADAKLNFIHKSNLDGTNRQAVVKGSLPHPFALTLFEDILYWT DWSTHSILACNKYTGEGLREIHSDIFSPMDIHAFSQQRQPNATNPCGIDNGGCSHLCLMS PVKPFYQCACPTGVKLLENGKTCKDGATELLLLARRTDLRRISLDTPDFTDIVLQLEDIR HAIAIDYDPVEGYIYWTDDEVRAIRRSFIDGSGSQFVVTAQIAHPDGIAVDWVARNLYWT DTGTDRIEVTRLNGTMRKILISEDLEEPRAIVLDPMVGYMYWTDWGEIPKIERAALDGSD RVVLVNTSLGWPNGLALDYDEGKIYWGDAKTDKIEVMNTDGTGRRVLVEDKIPHIFGFTL LGDYVYWTDWQRRSIERVHKRSAEREVIIDQLPDLMGLKATNVHRVIGSNPCAEENGGCS HLCLYRPQGLRCACPIGFELISDMKTCIVPEAFLLFSRRADIRRISLETNNNNVAIPLTG VKEASALDFDVTDNRIYWTDISLKTISRAFMNGSALEHVVEFGLDYPEGMAVDWLGKNLY WADTGTNRIEVSKLDGQHRQVLVWKDLDSPRALALDPAEGFMYWTEWGGKPKIDRAAMDG SERTTLVPNVGRANGLTIDYAKRRLYWTDLDTNLIESSNMLGLNREVIADDLPHPFGLTQ YQDYIYWTDWSRRSIERANKTSGQNRTIIQGHLDYVMDILVFHSSRQSGWNECASSNGHC SHLCLAVPVGGFVCGCPAHYSLNADNRTCSAPTTFLLFSQKSAINRMVIDEQQSPDIILP IHSLRNVRAIDYDPLDKQLYWIDSRQNMIRKAQEDGSQGFTVVVSSVPSQNLEIQPYDLS IDIYSRYIYWTCEATNVINVTRLDGRSVGVVLKGEQDRPRAVVVNPEKGYMYFTNLQERS PKIERAALDGTEREVLFFSGLSKPIALALDSRLGKLFWADSDLRRIESSDLSGANRIVLE DSNILQPVGLTVFENWLYWIDKQQQMIEKIDMTGREGRTKVQARIAQLSDIHAVKELNLQ EYRQHPCAQDNGGCSHICLVKGDGTTRCSCPMHLVLLQDELSCGEPPTCSPQQFTCFTGE IDCIPVAWRCDGFTECEDHSDELNCPVCSESQFQCASGQCIDGALRCNGDANCQDKSDEK NCEVLCLIDQFRCANGQCIGKHKKCDHNVDCSDKSDELDCYPTEEPAPQATNTVGSVIGV IVTIFVSGTVYFICQRMLCPRMKGDGETMTNDYVVHGPASVPLGYVPHPSSLSGSLPGMS RGKSMISSLSIMGGSSGPPYDRAHVTGASSSSSSSTKGTYFPAILNPPPSPATERSHYTM EFGYSSNSPSTHRSYSYRPYSYRHFAPPTTPCSTDVCDSDYAPSRRMTSVATAKGYTSDL NYDSEPVPPPPTPRSQYLSAEENYESCPPSPYTERSYSHHLYPPPPSPCTDSS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T02Z3O | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of GSK-3/Axin complex bound to phosphorylated Wnt receptor LRP6 e-motif | PDB:4NM7 | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | No | [3] |
| PDB Sequence |
PPPPC
|
|||||
|
|
||||||
| Ligand Name: Phosphonothreonine | Ligand Info | |||||
| Structure Description | Crystal structure of GSK-3/Axin complex bound to phosphorylated Wnt receptor LRP6 c-motif | PDB:4NM5 | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | No | [3] |
| PDB Sequence |
PPPPR
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| AXL receptor tyrosine kinase ligand (GAS6) | 50.000 (19/38) | 6.57E-05 | |
| Hemicentin-1 (HMCN1) | 26.667 (52/195) | 2.00E-03 |
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| mTOR signaling pathway | hsa04150 | Affiliated Target |
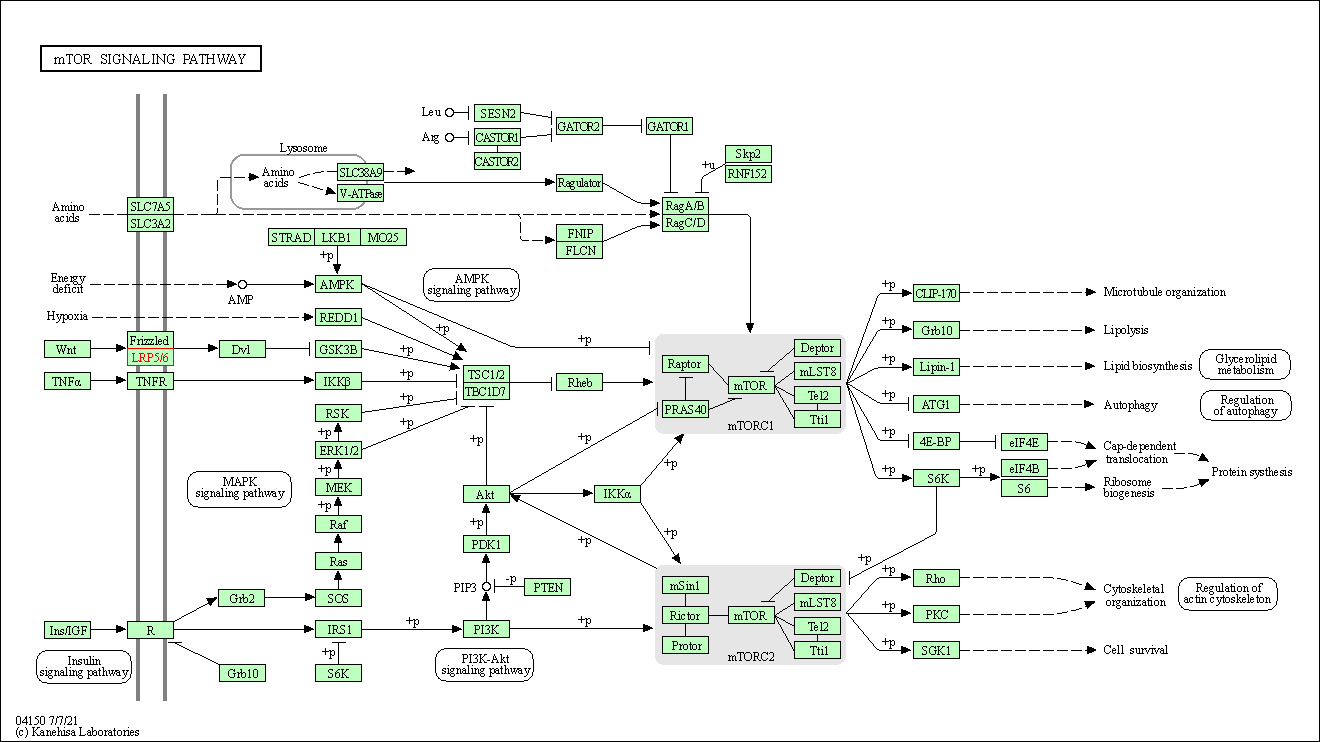
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Wnt signaling pathway | hsa04310 | Affiliated Target |
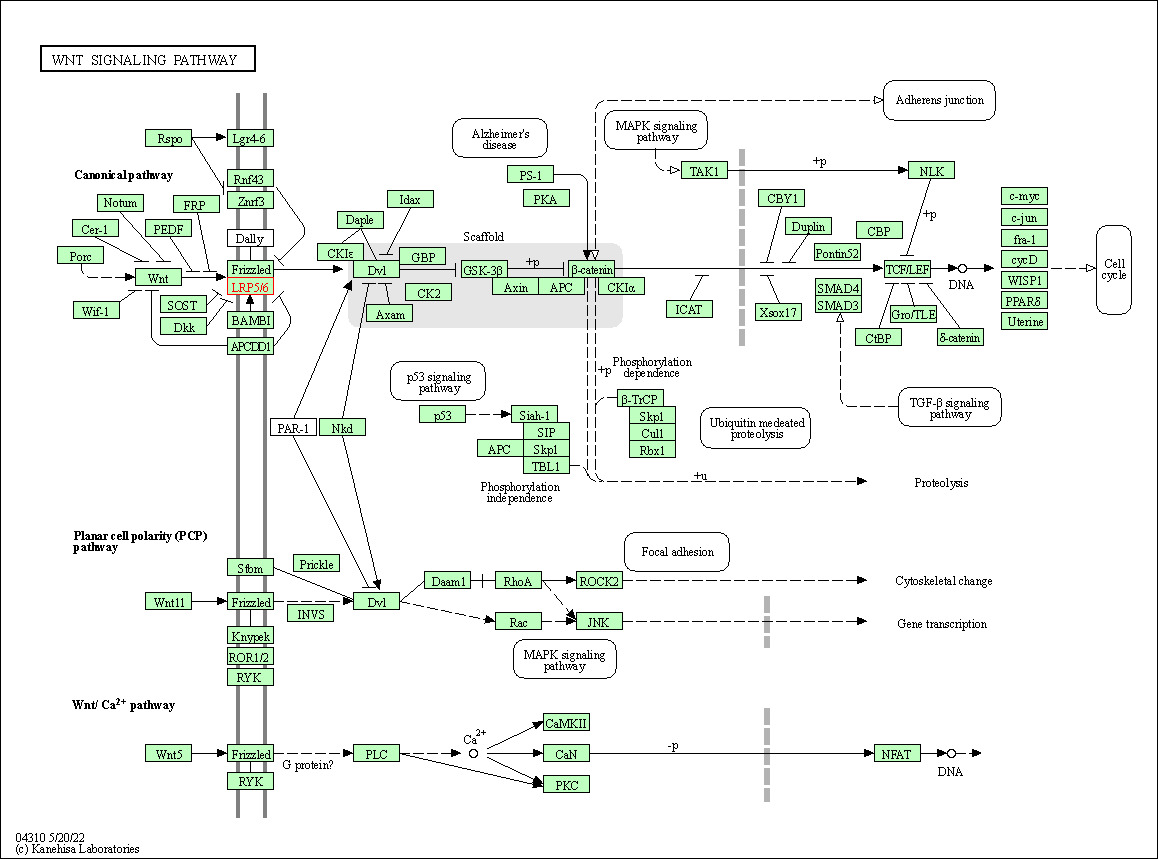
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |
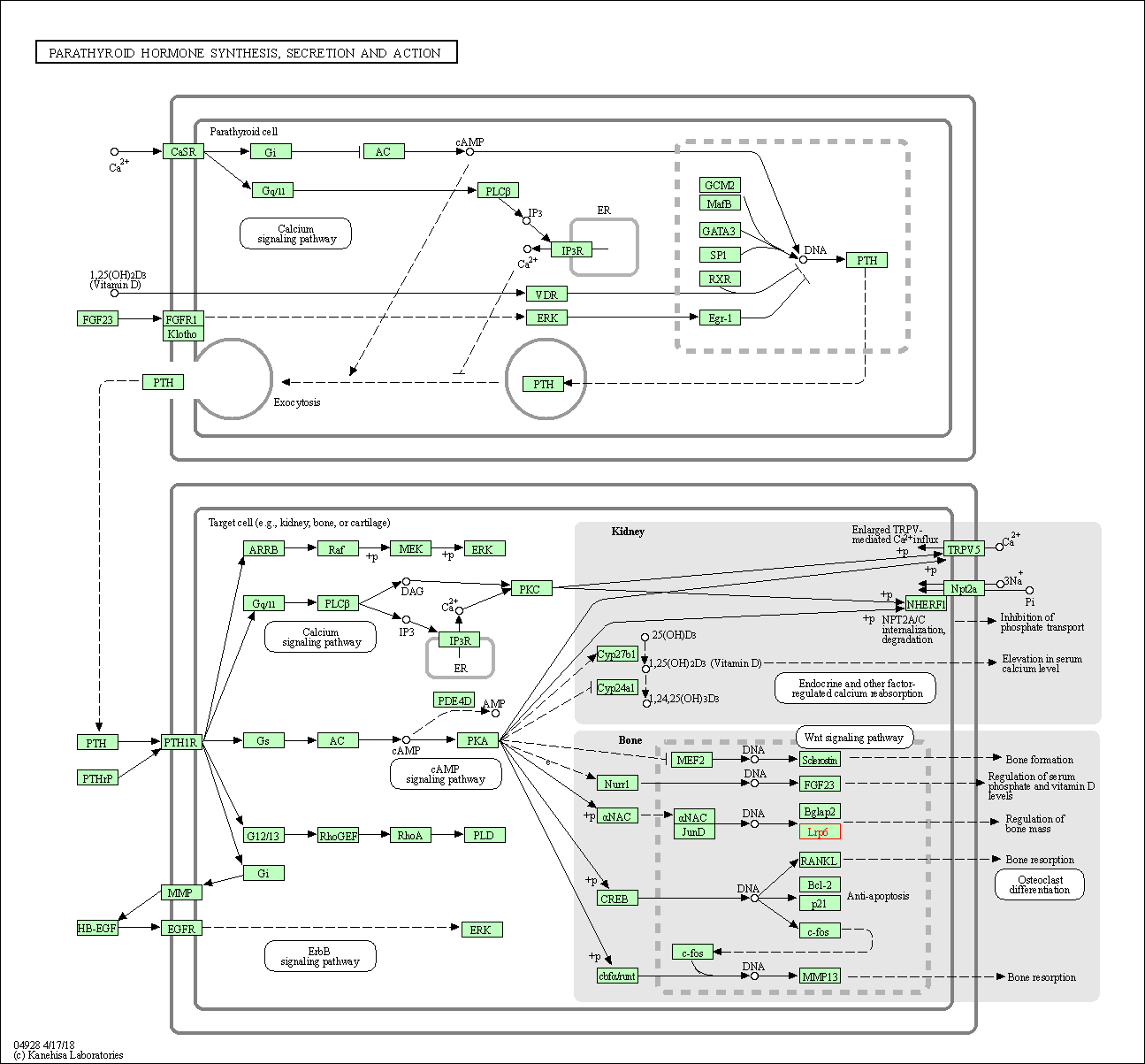
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 45 | Degree centrality | 4.83E-03 | Betweenness centrality | 3.36E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.40E-01 | Radiality | 1.42E+01 | Clustering coefficient | 1.23E-01 |
| Neighborhood connectivity | 1.86E+01 | Topological coefficient | 4.49E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Wnt signaling pathway | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Alzheimer disease-presenilin pathway | |||||
| 2 | Wnt signaling pathway | |||||
| PID Pathway | [+] 4 PID Pathways | + | ||||
| 1 | Degradation of beta catenin | |||||
| 2 | Presenilin action in Notch and Wnt signaling | |||||
| 3 | Wnt signaling network | |||||
| 4 | Canonical Wnt signaling pathway | |||||
| Reactome | [+] 4 Reactome Pathways | + | ||||
| 1 | TCF dependent signaling in response to WNT | |||||
| 2 | Negative regulation of TCF-dependent signaling by WNT ligand antagonists | |||||
| 3 | Disassembly of the destruction complex and recruitment of AXIN to the membrane | |||||
| 4 | Regulation of FZD by ubiquitination | |||||
| WikiPathways | [+] 6 WikiPathways | + | ||||
| 1 | Wnt Signaling Pathway | |||||
| 2 | Wnt Signaling Pathway and Pluripotency | |||||
| 3 | Wnt Signaling Pathway Netpath | |||||
| 4 | Hair Follicle Development: Induction (Part 1 of 3) | |||||
| 5 | Primary Focal Segmental Glomerulosclerosis FSGS | |||||
| 6 | MicroRNAs in cardiomyocyte hypertrophy | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991-5. | |||||
| REF 2 | ClinicalTrials.gov (NCT03604445) This Study Aims to Find and Test a Safe Dose of BI 905677 in Patients With Different Types of Cancer (Solid Tumours). U.S. National Institutes of Health. | |||||
| REF 3 | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6. Elife. 2014 Mar 18;3:e01998. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

