Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T51666
(Former ID: TTDS00044)
|
|||||
| Target Name |
Gastric H(+)/K(+) ATPase alpha (ATP4A)
|
|||||
| Synonyms |
Gastric H+/K+ ATPase alpha subunit; Gastric H(+)/K(+)-ATPase; ATP4A
Click to Show/Hide
|
|||||
| Gene Name |
ATP4A
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Bacterial infection [ICD-11: 1A00-1C4Z] | |||||
| 2 | Gastro-oesophageal reflux disease [ICD-11: DA22] | |||||
| 3 | Oesophageal ulcer [ICD-11: DA25] | |||||
| 4 | Peptic ulcer [ICD-11: DA61] | |||||
| Function |
Catalyzes the hydrolysis of ATP coupled with the exchange of H(+) and K(+) ions across the plasma membrane. Responsible for acid production in the stomach.
Click to Show/Hide
|
|||||
| BioChemical Class |
Acid anhydrides hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 7.2.2.19
|
|||||
| Sequence |
MGKAENYELYSVELGPGPGGDMAAKMSKKKKAGGGGGKRKEKLENMKKEMEINDHQLSVA
ELEQKYQTSATKGLSASLAAELLLRDGPNALRPPRGTPEYVKFARQLAGGLQCLMWVAAA ICLIAFAIQASEGDLTTDDNLYLAIALIAVVVVTGCFGYYQEFKSTNIIASFKNLVPQQA TVIRDGDKFQINADQLVVGDLVEMKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTRSP ECTHESPLETRNIAFFSTMCLEGTVQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIE HFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVTVCLSLTAK RLASKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHIHTADTTEDQSGQ TFDQSSETWRALCRVLTLCNRAAFKSGQDAVPVPKRIVIGDASETALLKFSELTLGNAMG YRDRFPKVCEIPFNSTNKFQLSIHTLEDPRDPRHLLVMKGAPERVLERCSSILIKGQELP LDEQWREAFQTAYLSLGGLGERVLGFCQLYLNEKDYPPGYAFDVEAMNFPSSGLCFAGLV SMIDPPRATVPDAVLKCRTAGIRVIMVTGDHPITAKAIAASVGIISEGSETVEDIAARLR VPVDQVNRKDARACVINGMQLKDMDPSELVEALRTHPEMVFARTSPQQKLVIVESCQRLG AIVAVTGDGVNDSPALKKADIGVAMGIAGSDAAKNAADMILLDDNFASIVTGVEQGRLIF DNLKKSIAYTLTKNIPELTPYLIYITVSVPLPLGCITILFIELCTDIFPSVSLAYEKAES DIMHLRPRNPKRDRLVNEPLAAYSYFQIGAIQSFAGFTDYFTAMAQEGWFPLLCVGLRAQ WEDHHLQDLQDSYGQEWTFGQRLYQQYTCYTVFFISIEVCQIADVLIRKTRRLSAFQQGF FRNKILVIAIVFQVCIGCFLCYCPGMPNIFNFMPIRFQWWLVPLPYGILIFVYDEIRKLG VRCCPGSWWDQELYY Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T68J49 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Dexlansoprazole | Drug Info | Approved | Non-erosive gastro-esophageal reflux disease | [2] | |
| 2 | Rabeprazole | Drug Info | Approved | Bacterial infection | [3], [4] | |
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Esomeprazole magnesium + Aspirin | Drug Info | Phase 3 | Peptic ulcer | [4] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | Saliphenylhalamide | Drug Info | Preclinical | Solid tumour/cancer | [7] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Dexlansoprazole | Drug Info | [8] | |||
| 2 | Rabeprazole | Drug Info | [1], [9] | |||
| 3 | Esomeprazole magnesium + Aspirin | Drug Info | [10] | |||
| 4 | Saliphenylhalamide | Drug Info | [11] | |||
| 5 | CDRI-85/92 | Drug Info | [11] | |||
| Modulator | [+] 4 Modulator drugs | + | ||||
| 1 | ARH-1029 | Drug Info | [11] | |||
| 2 | CA-50040 | Drug Info | [11] | |||
| 3 | OPC-22575 | Drug Info | [11] | |||
| 4 | PA-10040 | Drug Info | [11] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Oxidative phosphorylation | hsa00190 | Affiliated Target |
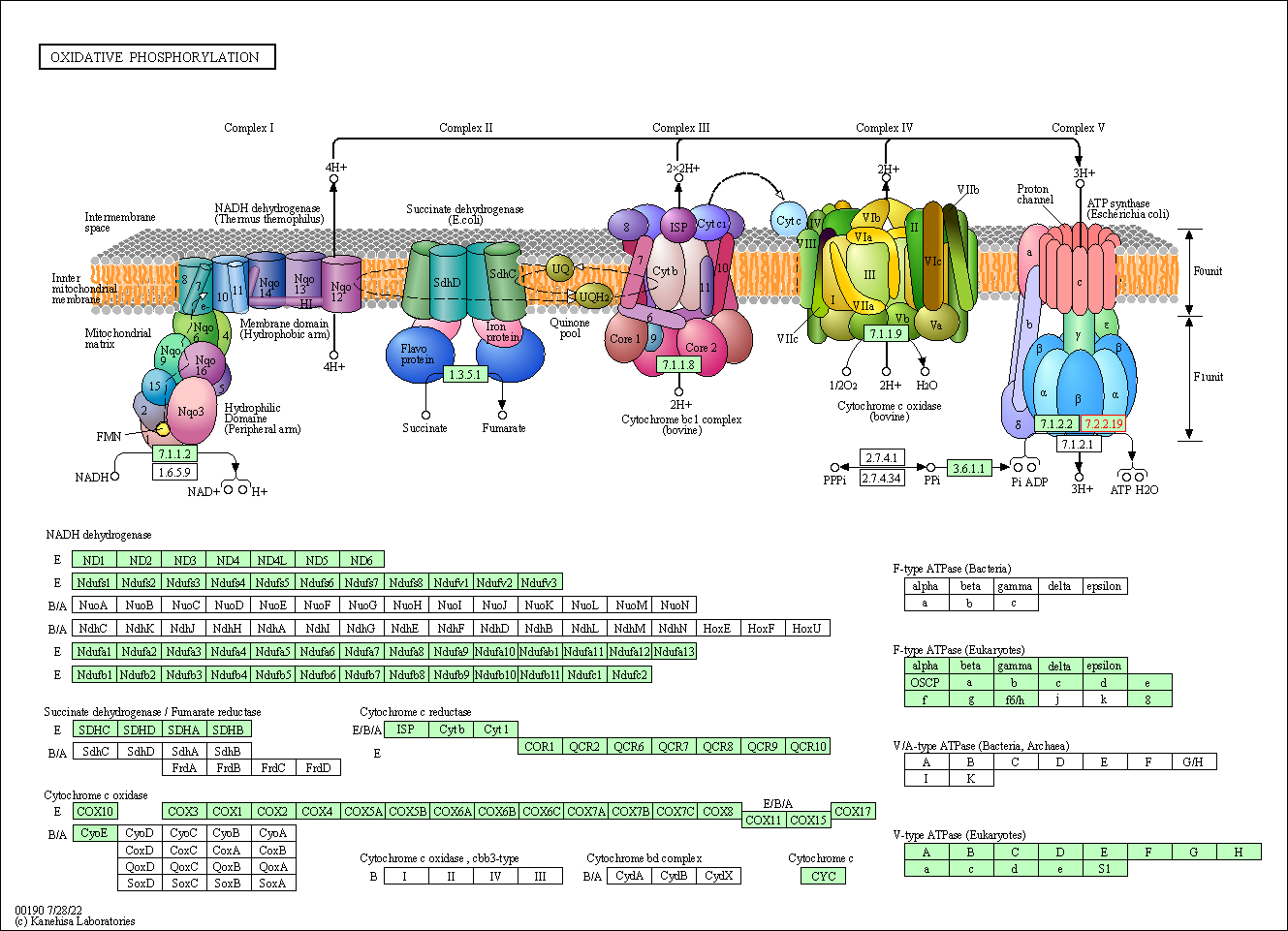
|
| Class: Metabolism => Energy metabolism | Pathway Hierarchy | ||
| Collecting duct acid secretion | hsa04966 | Affiliated Target |

|
| Class: Organismal Systems => Excretory system | Pathway Hierarchy | ||
| Gastric acid secretion | hsa04971 | Affiliated Target |
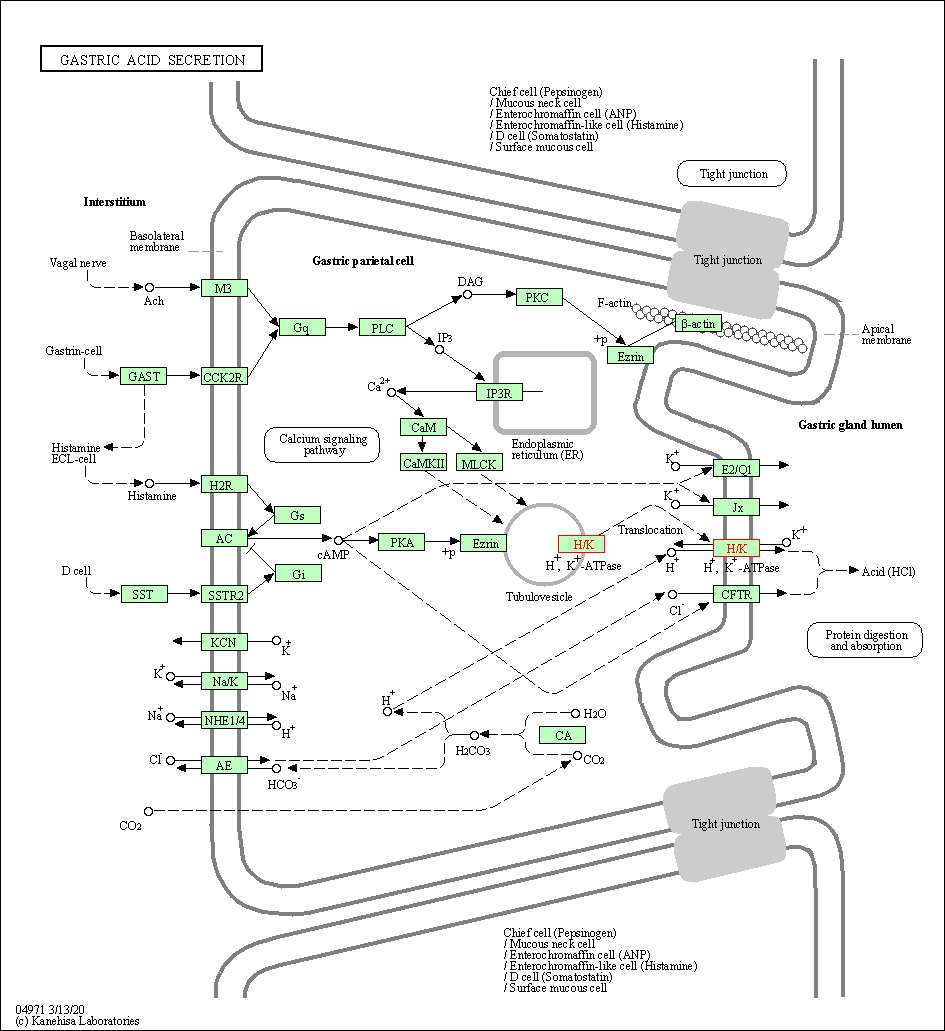
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 1.01E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.43E-01 | Radiality | 1.17E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 2.50E+00 | Topological coefficient | 7.50E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Oxidative phosphorylation | |||||
| 2 | Collecting duct acid secretion | |||||
| 3 | Gastric acid secretion | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Gastric Acid Production | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Ion transport by P-type ATPases | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | Iron uptake and transport | |||||
| 2 | Secretion of Hydrochloric Acid in Parietal Cells | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Review article: rabeprazole's tolerability profile in clinical trials. Aliment Pharmacol Ther. 1999 Oct;13 Suppl 5:17-23. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 022287. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7290). | |||||
| REF 4 | An overview of proton pump inhibitors. Gastroenterol Nurs. 2003 Sep-Oct;26(5):182-90. | |||||
| REF 5 | ClinicalTrials.gov (NCT01400945) Efficacy Study of AGSPT201 Tablet to Treat Gastroesophageal Reflux Disease. U.S. National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT00325715) AGN 201904 Versus Esomeprazole in the Prevention of Aspirin-induced Stomach or Upper Intestinal Damage in Healthy Volunteers. U.S. National Institutes of Health. | |||||
| REF 7 | Lysosomes as a therapeutic target. Nat Rev Drug Discov. 2019 Dec;18(12):923-948. | |||||
| REF 8 | Clinical pipeline report, company report or official report of Takeda (2009). | |||||
| REF 9 | Review article: pharmacokinetic concerns in the selection of anti-ulcer therapy. Aliment Pharmacol Ther. 1999 Oct;13 Suppl 5:11-6. | |||||
| REF 10 | Clinical pipeline report, company report or official report of AstraZeneca (2009). | |||||
| REF 11 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 849). | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

