Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T59230
(Former ID: TTDR00811)
|
|||||
| Target Name |
Estradiol 17 beta-dehydrogenase 4 (HSD17B4)
|
|||||
| Synonyms |
MFE-2; HSD17B4; DBP; D-bifunctional protein; 17-beta-hydroxysteroid dehydrogenase 4; 17-beta-HSD 4; 17 beta-hydroxysteroid dehydrogenase type 4
Click to Show/Hide
|
|||||
| Gene Name |
HSD17B4
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Bifunctional enzymeacting on the peroxisomal beta- oxidation pathway for fatty acids. Catalyzes the formation of 3- ketoacyl-CoA intermediates from both straight-chain and 2-methyl- branched-chain fatty acids.
Click to Show/Hide
|
|||||
| BioChemical Class |
Short-chain dehydrogenases reductase
|
|||||
| UniProt ID | ||||||
| Sequence |
MGSPLRFDGRVVLVTGAGAGLGRAYALAFAERGALVVVNDLGGDFKGVGKGSLAADKVVE
EIRRRGGKAVANYDSVEEGEKVVKTALDAFGRIDVVVNNAGILRDRSFARISDEDWDIIH RVHLRGSFQVTRAAWEHMKKQKYGRIIMTSSASGIYGNFGQANYSAAKLGLLGLANSLAI EGRKSNIHCNTIAPNAGSRMTQTVMPEDLVEALKPEYVAPLVLWLCHESCEENGGLFEVG AGWIGKLRWERTLGAIVRQKNHPMTPEAVKANWKKICDFENASKPQSIQESTGSIIEVLS KIDSEGGVSANHTSRATSTATSGFAGAIGQKLPPFSYAYTELEAIMYALGVGASIKDPKD LKFIYEGSSDFSCLPTFGVIIGQKSMMGGGLAEIPGLSINFAKVLHGEQYLELYKPLPRA GKLKCEAVVADVLDKGSGVVIIMDVYSYSEKELICHNQFSLFLVGSGGFGGKRTSDKVKV AVAIPNRPPDAVLTDTTSLNQAALYRLSGDWNPLHIDPNFASLAGFDKPILHGLCTFGFS ARRVLQQFADNDVSRFKAIKARFAKPVYPGQTLQTEMWKEGNRIHFQTKVQETGDIVISN AYVDLAPTSGTSAKTPSEGGKLQSTFVFEEIGRRLKDIGPEVVKKVNAVFEWHITKGGNI GAKWTIDLKSGSGKVYQGPAKGAADTTIILSDEDFMEVVLGKLDPQKAFFSGRLKARGNI MLSQKLQMILKDYAKL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Nicotinamide-Adenine-Dinucleotide | Ligand Info | |||||
| Structure Description | Crystal Structure Of Human 17-Beta-Hydroxysteroid Dehydrogenase Type 4 In Complex With NAD | PDB:1ZBQ | ||||
| Method | X-ray diffraction | Resolution | 2.19 Å | Mutation | No | [3] |
| PDB Sequence |
SPLRFDGRVV
12 LVTGAGAGLG22 RAYALAFAER32 GALVVVNDLG42 GDFKGVGKGS52 LAADKVVEEI 62 RRRGGKAVAN72 YDSVEEGEKV82 VKTALDAFGR92 IDVVVNNAGI102 LRDRSFARIS 112 DEDWDIIHRV122 HLRGSFQVTR132 AAWEHMKKQK142 YGRIIMTSSA152 SGIYGNFGQA 162 NYSAAKLGLL172 GLANSLAIEG182 RKSNIHCNTI192 APNAGSRMTQ202 TVMPEDLVEA 212 LKPEYVAPLV222 LWLCHESCEE232 NGGLFEVGAG242 WIGKLRWERT252 LGAIVRQKNH 262 PMTPEAVKAN272 WKKICDFENA282 SKPQSIQEST292 GSIIEVLSKI302 DS |
|||||
|
|
THR15
4.955
GLY16
3.403
ALA17
4.987
GLY18
4.070
ALA19
3.589
GLY20
3.469
LEU21
2.861
GLY22
4.436
ASN39
3.288
ASP40
2.406
LEU41
3.494
GLY42
4.023
GLY43
3.897
PHE45
4.255
ASP74
4.032
SER75
2.730
VAL76
3.158
GLU77
4.417
ASN99
2.878
|
|||||
| Ligand Name: Oxtoxynol-10 | Ligand Info | |||||
| Structure Description | LIGANDED STEROL CARRIER PROTEIN TYPE 2 (SCP-2) LIKE DOMAIN OF HUMAN MULTIFUNCTIONAL ENZYME TYPE 2 (MFE-2) | PDB:1IKT | ||||
| Method | X-ray diffraction | Resolution | 1.75 Å | Mutation | No | [4] |
| PDB Sequence |
LQSTFVFEEI
15 GRRLKDIGPE25 VVKKVNAVFE35 WHITKGGNIG45 AKWTIDLKSG55 SGKVYQGPAK 65 GAADTTIILS75 DEDFMEVVLG85 KLDPQKAFFS95 GRLKARGNIM105 LSQKLQMILK 115 DYAKL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Primary bile acid biosynthesis | hsa00120 | Affiliated Target |
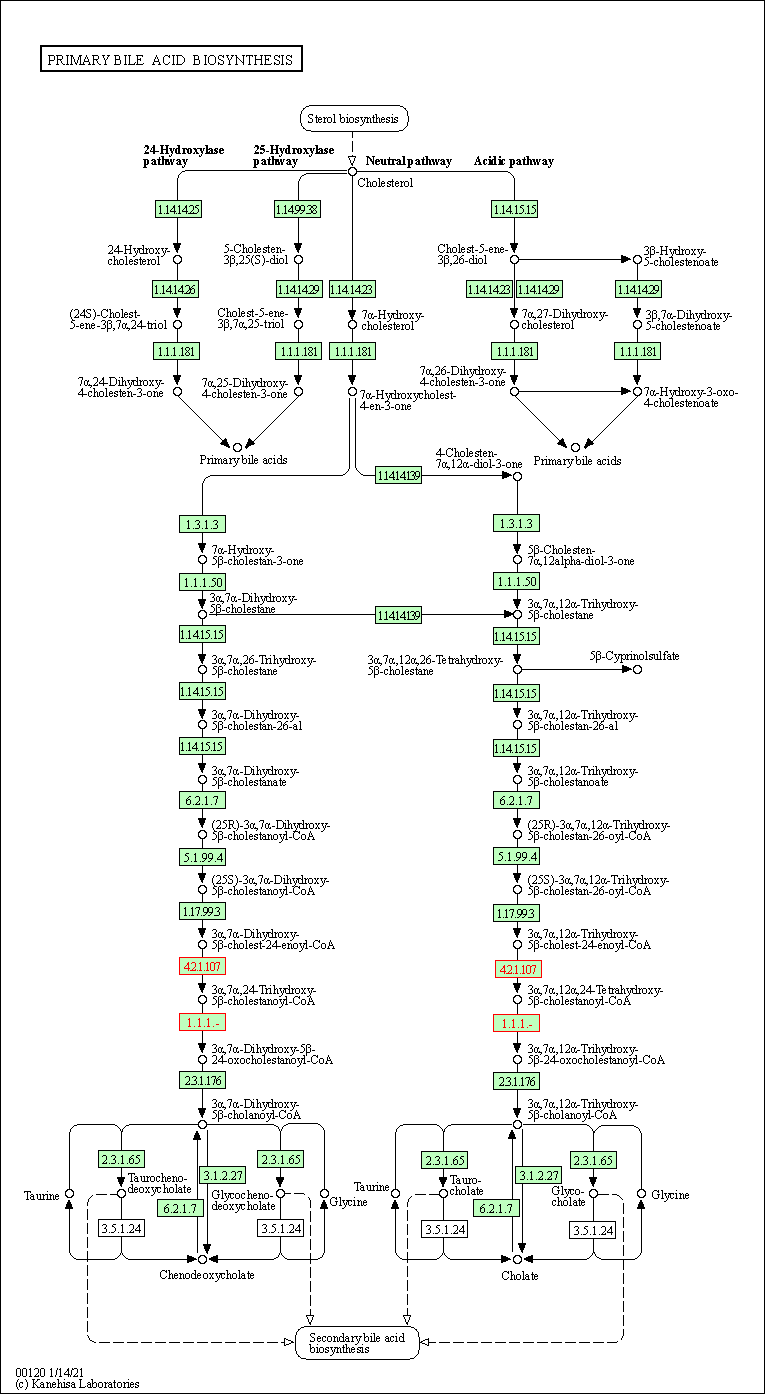
|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
| Biosynthesis of unsaturated fatty acids | hsa01040 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
| Peroxisome | hsa04146 | Affiliated Target |
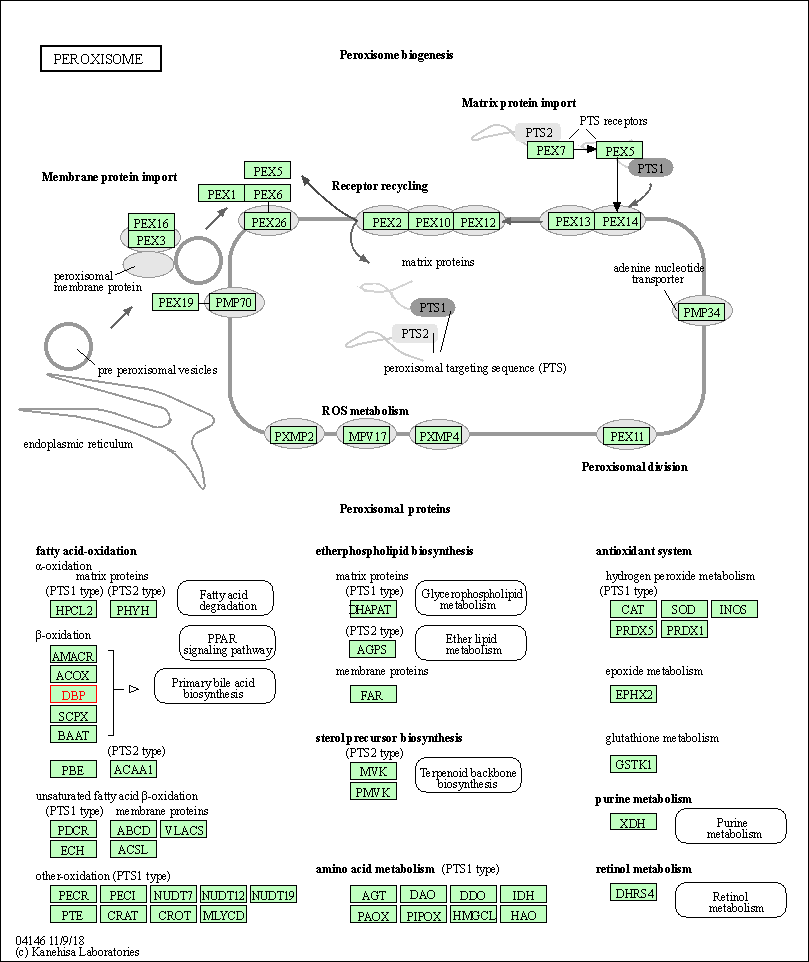
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Degree | 16 | Degree centrality | 1.72E-03 | Betweenness centrality | 1.70E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.95E-01 | Radiality | 1.33E+01 | Clustering coefficient | 5.67E-01 |
| Neighborhood connectivity | 2.10E+01 | Topological coefficient | 2.23E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 1 BioCyc Pathways | + | ||||
| 1 | Fatty acid beta-oxidation (peroxisome) | |||||
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Primary bile acid biosynthesis | |||||
| 2 | Metabolic pathways | |||||
| 3 | Peroxisome | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Bile Acid Biosynthesis | |||||
| WikiPathways | [+] 6 WikiPathways | + | ||||
| 1 | Steroid Biosynthesis | |||||
| 2 | alpha-linolenic (omega3) and linoleic (omega6) acid metabolism | |||||
| 3 | Prostate Cancer | |||||
| 4 | Peroxisomal beta-oxidation of tetracosanoyl-CoA | |||||
| 5 | Peroxisomal lipid metabolism | |||||
| 6 | Bile acid and bile salt metabolism | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 2 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 3 | Crystal Structure Of Human 17-Beta-Hydroxysteroid Dehydrogenase Type 4 In Complex With NAD | |||||
| REF 4 | Crystal structure of the liganded SCP-2-like domain of human peroxisomal multifunctional enzyme type 2 at 1.75 A resolution. J Mol Biol. 2001 Nov 9;313(5):1127-38. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

