Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T62193
(Former ID: TTDS00361)
|
|||||
| Target Name |
Aromatic-L-amino-acid decarboxylase (DDC)
|
|||||
| Synonyms |
DOPA decarboxylase; AADC
Click to Show/Hide
|
|||||
| Gene Name |
DDC
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Parkinsonism [ICD-11: 8A00] | |||||
| 2 | Vitamin deficiency [ICD-11: 5B55-5B5F] | |||||
| Function |
Catalyzes the decarboxylation of L-3,4-dihydroxyphenylalanine (DOPA) to dopamine, L-5-hydroxytryptophan to serotonin and L-tryptophan to tryptamine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Carbon-carbon lyase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 4.1.1.28
|
|||||
| Sequence |
MNASEFRRRGKEMVDYMANYMEGIEGRQVYPDVEPGYLRPLIPAAAPQEPDTFEDIINDV
EKIIMPGVTHWHSPYFFAYFPTASSYPAMLADMLCGAIGCIGFSWAASPACTELETVMMD WLGKMLELPKAFLNEKAGEGGGVIQGSASEATLVALLAARTKVIHRLQAASPELTQAAIM EKLVAYSSDQAHSSVERAGLIGGVKLKAIPSDGNFAMRASALQEALERDKAAGLIPFFMV ATLGTTTCCSFDNLLEVGPICNKEDIWLHVDAAYAGSAFICPEFRHLLNGVEFADSFNFN PHKWLLVNFDCSAMWVKKRTDLTGAFRLDPTYLKHSHQDSGLITDYRHWQIPLGRRFRSL KMWFVFRMYGVKGLQAYIRKHVQLSHEFESLVRQDPRFEICVEVILGLVCFRLKGSNKVN EALLQRINSAKKIHLVPCHLRDKFVLRFAICSRTVESAHVQRAWEHIKELAADVLRAERE Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A01759 | |||||
| HIT2.0 ID | T40TBX | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Carbidopa | Drug Info | Approved | Parkinson disease | [2], [3] | |
| 2 | Vitamin B6 | Drug Info | Approved | Vitamin B6 deficiency | [4], [5] | |
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | Patrome | Drug Info | Phase 3 | Parkinson disease | [6] | |
| 2 | AV-201 | Drug Info | Phase 2 | Parkinson disease | [10] | |
| 3 | ProSavin | Drug Info | Phase 1/2 | Parkinson disease | [12] | |
| Mode of Action | [+] 4 Modes of Action | + | ||||
| Inhibitor | [+] 2 Inhibitor drugs | + | ||||
| 1 | Carbidopa | Drug Info | [14] | |||
| 2 | 3-hydroxybenzylhydrazine | Drug Info | [18] | |||
| Cofactor | [+] 1 Cofactor drugs | + | ||||
| 1 | Vitamin B6 | Drug Info | [1] | |||
| Modulator | [+] 3 Modulator drugs | + | ||||
| 1 | Patrome | Drug Info | [15] | |||
| 2 | AV-201 | Drug Info | [16] | |||
| 3 | ProSavin | Drug Info | [17] | |||
| Activator | [+] 1 Activator drugs | + | ||||
| 1 | Noradrenalone (arterenone) | Drug Info | [19] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Pyridoxal phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the open conformation with LLP and PLP bound to Chain-A and Chain-B respectively | PDB:3RCH | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [20] |
| PDB Sequence |
MNASEFRRRG
10 KEMVDYVANY20 MEGIEGRQVY30 PDVEPGYLRP40 LIPAAAPQEP50 DTFEDIINDV 60 EKIIMPGVTH70 WHSPYFFAYF80 PTASSYPAML90 ADMLCGAIGA107 SPACTELETV 117 MMDWLGKMLE127 LPKAFLNEKA137 GEGGGVIQGS147 ASEATLVALL157 AARTKVIHRL 167 QAASPELTQA177 AIMEKLVAYS187 SDQAHSSVER197 AGLIGGVKLK207 AIPSDGNFAM 217 RASALQEALE227 RDKAAGLIPF237 FMVATLGTTT247 CCSFDNLLEV257 GPICNKEDIW 267 LHVDAAYAGS277 AFICPEFRHL287 LNGVEFADSF297 NFNPHKWLLV307 NFDCSAMWVK 317 KRTDLRFRSL360 KMWFVFRMYG370 VKGLQAYIRK380 HVQLSHEFES390 LVRQDPRFEI 400 CVEVILGLVC410 FRLKGSNKVN420 EALLQRINSA430 KKIHLVPCHL440 RDKFVLRFAI 450 CSRTVESAHV460 QRAWEHIKEL470 AADVLRAERE480
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: N6-((3-Hydroxy-2-methyl-5-((phosphonooxy)methyl)-4-pyridinyl)methylene)-L-lysine | Ligand Info | |||||
| Structure Description | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the open conformation with LLP and PLP bound to Chain-A and Chain-B respectively | PDB:3RCH | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [20] |
| PDB Sequence |
MNASEFRRRG
10 KEMVDYVANY20 MEGIEGRQVY30 PDVEPGYLRP40 LIPAAAPQEP50 DTFEDIINDV 60 EKIIMPGVTH70 WHSPYFFAYF80 PTASSYPAML90 ADMLCGAIGC100 ISPACTELET 116 VMMDWLGKML126 ELPKAFLNEK136 AGEGGGVIQG146 SASEATLVAL156 LAARTKVIHR 166 LQAASPELTQ176 AAIMEKLVAY186 SSDQAHSSVE196 RAGLIGGVKL206 KAIPSDGNFA 216 MRASALQEAL226 ERDKAAGLIP236 FFMVATLGTT246 TCCSFDNLLE256 VGPICNKEDI 266 WLHVDAAYAG276 SAFICPEFRH286 LLNGVEFADS296 FNFNPHWLLV307 NFDCSAMWVK 317 KRTDLTGAFR355 RFRSLKMWFV365 FRMYGVKGLQ375 AYIRKHVQLS385 HEFESLVRQD 395 PRFEICVEVI405 LGLVCFRLKG415 SNKVNEALLQ425 RINSAKKIHL435 VPCHLRDKFV 445 LRFAICSRTV455 ESAHVQRAWE465 HIKELAADVL475 RAERE
|
|||||
|
|
TYR79
3.051
PHE80
3.991
PRO81
2.468
THR82
3.654
ALA83
3.401
GLY146
4.747
SER147
3.104
ALA148
2.968
SER149
2.794
HIS192
3.241
SER194
4.163
THR242
3.838
GLY244
3.724
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Tyrosine metabolism | hsa00350 | Affiliated Target |
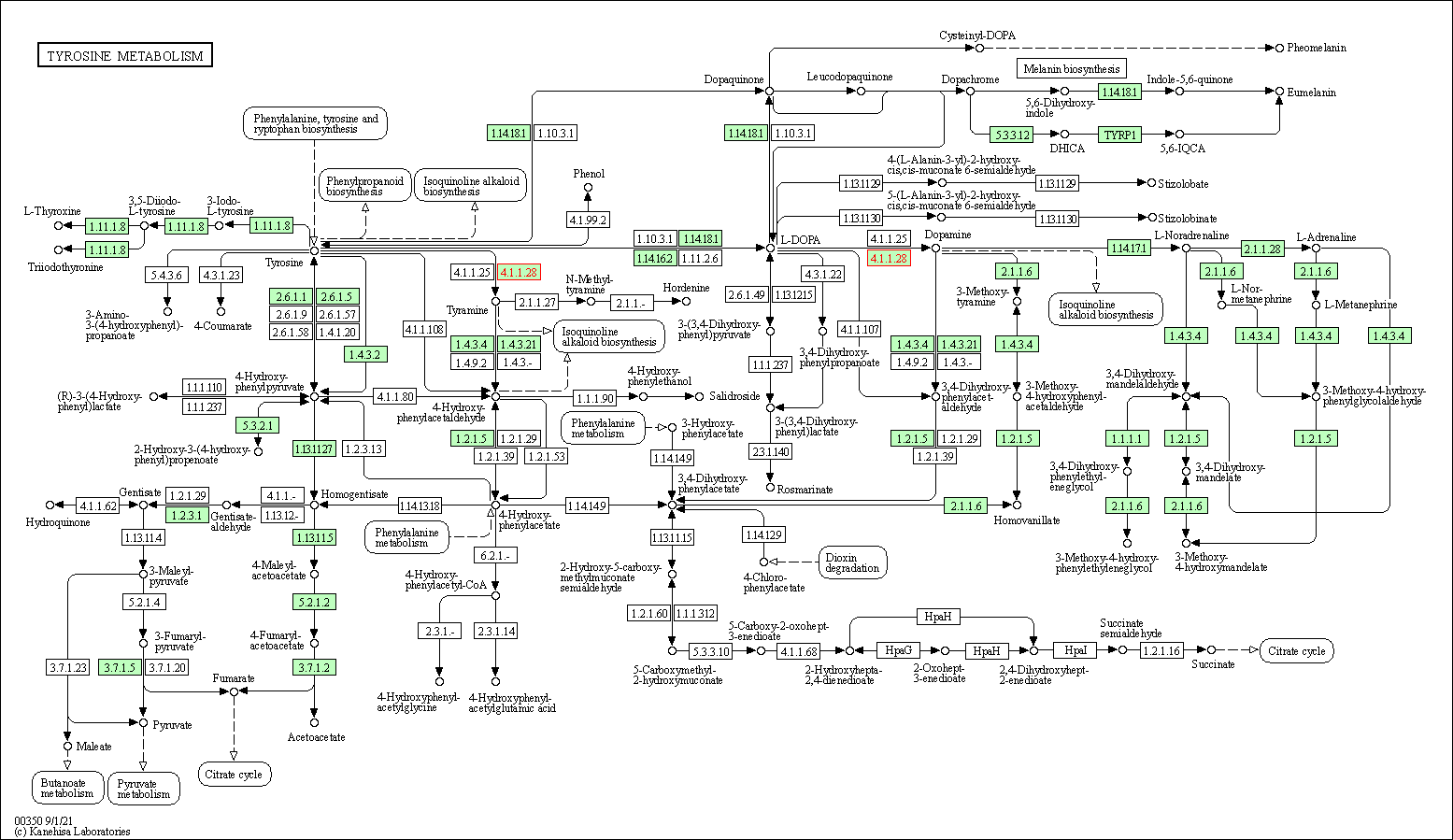
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Phenylalanine metabolism | hsa00360 | Affiliated Target |
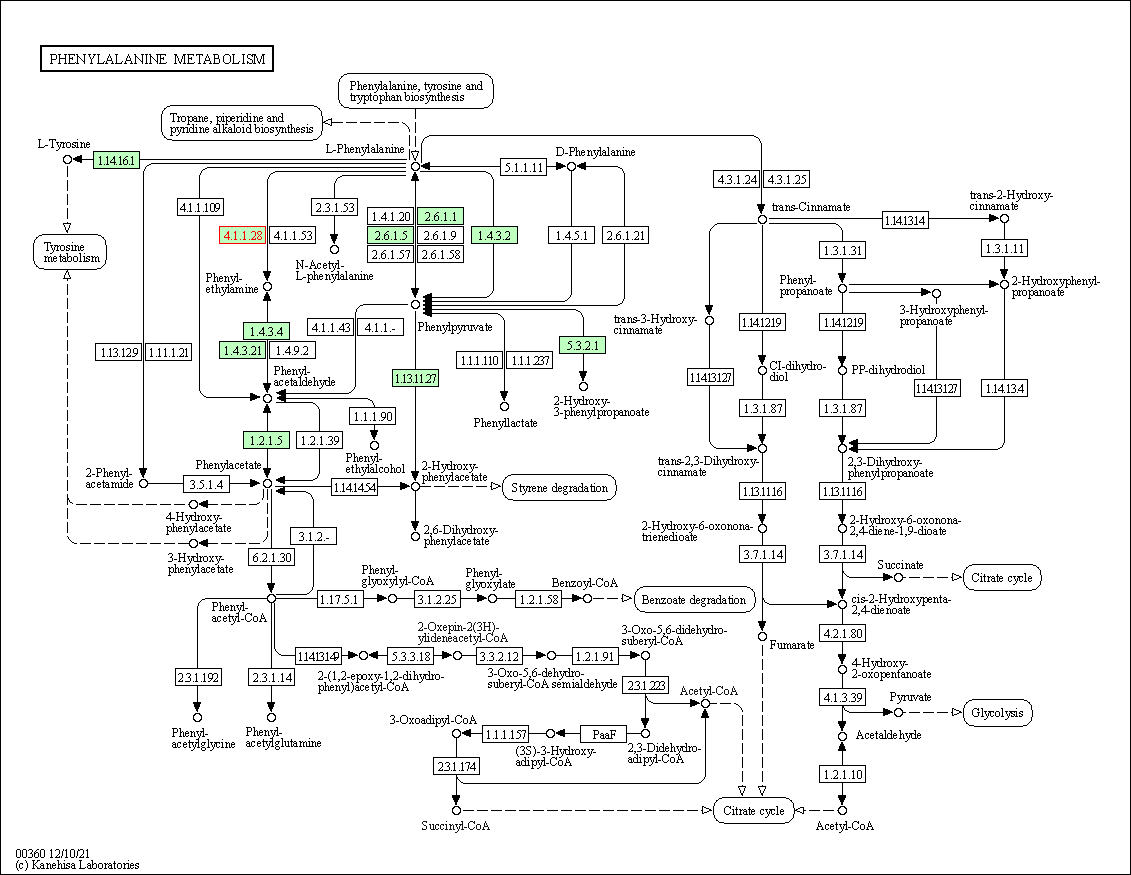
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Tryptophan metabolism | hsa00380 | Affiliated Target |
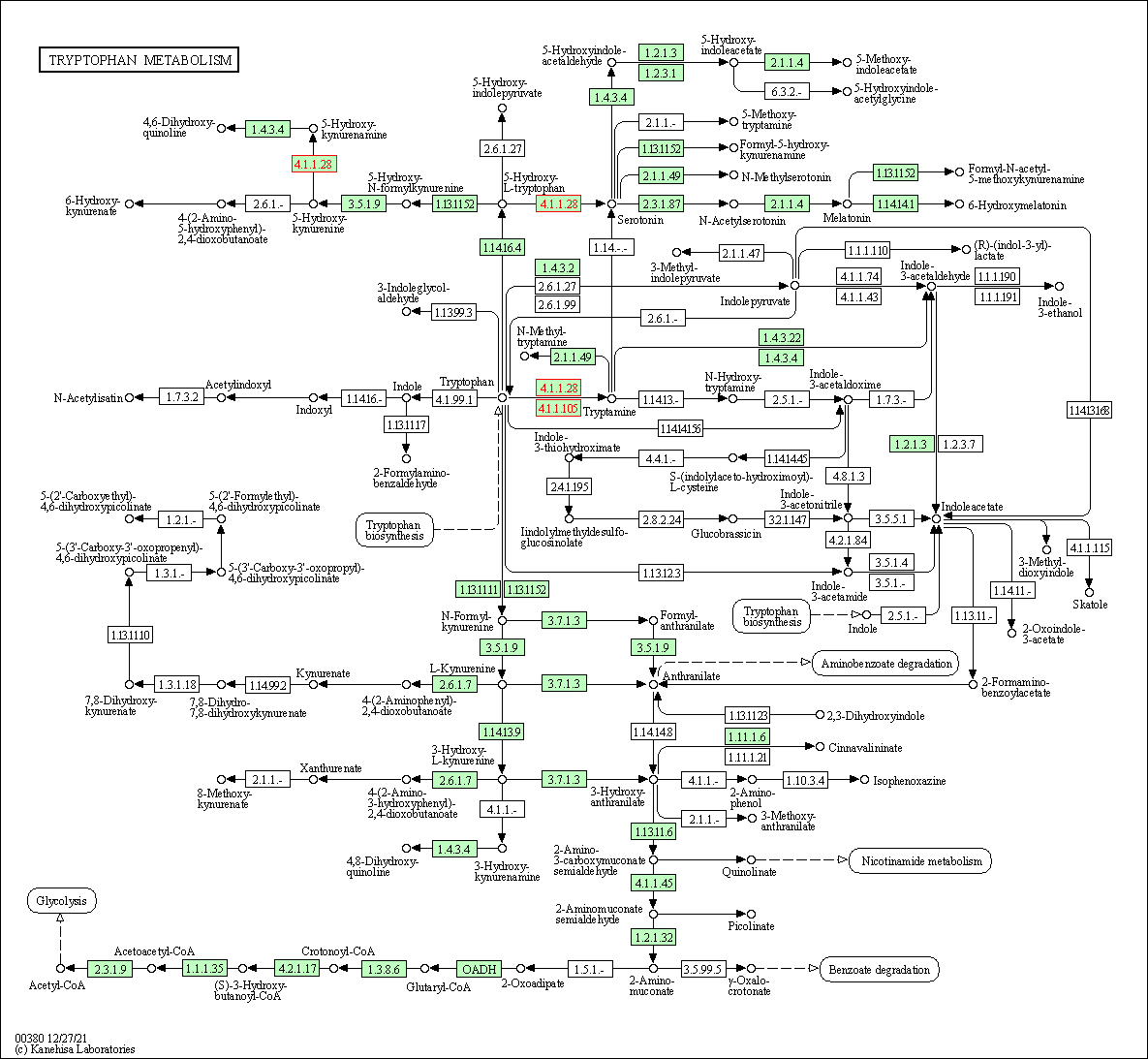
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Serotonergic synapse | hsa04726 | Affiliated Target |
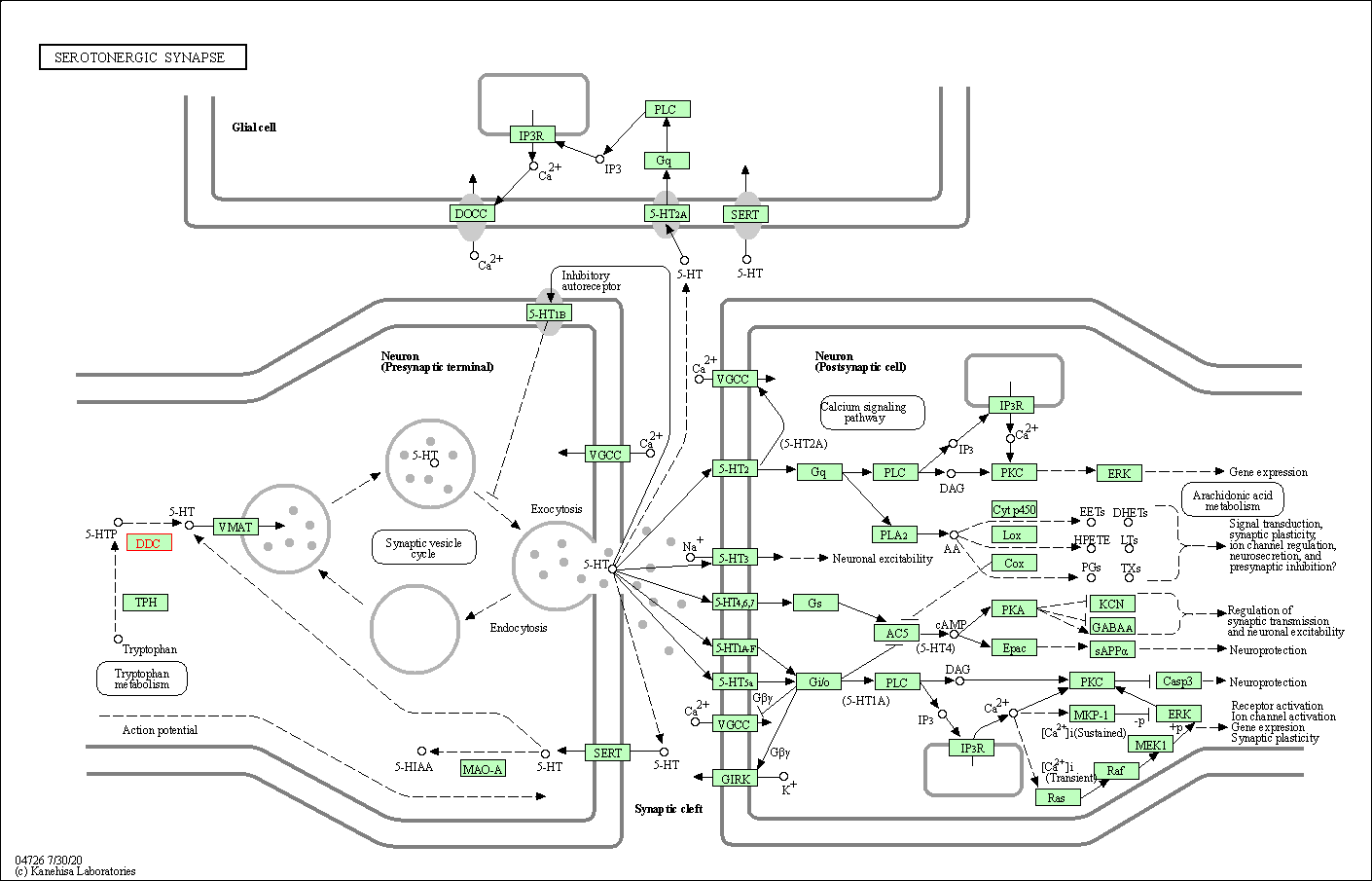
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Degree | 11 | Degree centrality | 1.18E-03 | Betweenness centrality | 1.05E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 1.78E-01 | Radiality | 1.29E+01 | Clustering coefficient | 7.27E-02 |
| Neighborhood connectivity | 4.00E+00 | Topological coefficient | 1.53E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Functional COMT variant predicts response to high dose pyridoxine in Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet. 2005 Aug 5;137B(1):1-4. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5159). | |||||
| REF 3 | Emerging drugs for restless legs syndrome. Expert Opin Emerg Drugs. 2005 Aug;10(3):537-52. | |||||
| REF 4 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 5 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 010598. | |||||
| REF 6 | Dopamine dysregulation syndrome, addiction and behavioral changes in Parkinson's disease. Parkinsonism Relat Disord. 2008;14(4):273-80. | |||||
| REF 7 | Clinical pipeline report, company report or official report of PTC Therapeutics. | |||||
| REF 8 | ClinicalTrials.gov (NCT03562494) VY-AADC02 for Parkinson's Disease With Motor Fluctuations (RESTORE-1). U.S. National Institutes of Health. | |||||
| REF 9 | ClinicalTrials.gov (NCT04903288) An Open-Label Trial to Address the Safety of the SmartFlow MR-Compatible Ventricular Cannula for Administering Eladocagene Exuparvovec to Pediatric Subjects. U.S.National Institutes of Health. | |||||
| REF 10 | Clinical pipeline report, company report or official report of Genzyme. | |||||
| REF 11 | ClinicalTrials.gov (NCT03720418) Study of OXB-102 (AXO-Lenti-PD) in Patients With Bilateral, Idiopathic Parkinson's Disease (SUNRISE-PD). U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT01856439) Long Term Safety and Efficacy Study of ProSavin in Parkinson's Disease. U.S. National Institutes of Health. | |||||
| REF 13 | ClinicalTrials.gov (NCT03065192) An Open-label Safety and Efficacy Study of VY-AADC01 Administered by MRI-Guided Convective Infusion Using a Posterior Trajectory Into the Putamen of Participants With Parkinson's Disease With Fluctuating Responses to Levodopa. U.S.National Institutes of Health. | |||||
| REF 14 | Catechol-O-methyltransferase inhibitors in the management of Parkinson's disease. Semin Neurol. 2001;21(1):15-22. | |||||
| REF 15 | The aromatic-L-amino acid decarboxylase inhibitor carbidopa is selectively cytotoxic to human pulmonary carcinoid and small cell lung carcinoma cells. Clin Cancer Res. 2000 Nov;6(11):4365-72. | |||||
| REF 16 | UCSF report | |||||
| REF 17 | Clinical pipeline report, company report or official report of Oxford BioMedica. | |||||
| REF 18 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1271). | |||||
| REF 19 | Activation of an adrenergic pro-drug through sequential stereoselective action of tandem target enzymes. Biochem Biophys Res Commun. 1992 Nov 30;189(1):33-9. | |||||
| REF 20 | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20514-9. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

