Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T66161
(Former ID: TTDR00385)
|
|||||
| Target Name |
Filamin A (FLNA)
|
|||||
| Synonyms |
Nonmuscle filamin; Non-muscle filamin; Filamin-A; Filamin-1; Filamin 1; FLN1; FLN-A; FLN; Endothelial actin-binding protein; Endothelial actin cytoskeleton; Alpha-filamin; Actin-binding protein 280; ABP-280
Click to Show/Hide
|
|||||
| Gene Name |
FLNA
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Alzheimer disease [ICD-11: 8A20] | |||||
| Function |
Anchors various transmembrane proteins to the actin cytoskeleton and serves as a scaffold for a wide range of cytoplasmic signaling proteins. Interaction with FLNA may allow neuroblast migration from the ventricular zone into the cortical plate. Tethers cell surface-localized furin, modulates its rate of internalization and directs its intracellular trafficking. Involved in ciliogenesis. Plays a role in cell-cell contacts and adherens junctions during the development of blood vessels, heart and brain organs. Plays a role in platelets morphology through interaction with SYK that regulates ITAM- and ITAM-like-containing receptor signaling, resulting in by platelet cytoskeleton organization maintenance. Promotes orthogonal branching of actin filaments and links actin filaments to membrane glycoproteins.
Click to Show/Hide
|
|||||
| BioChemical Class |
Dystrophin protein
|
|||||
| UniProt ID | ||||||
| Sequence |
MSSSHSRAGQSAAGAAPGGGVDTRDAEMPATEKDLAEDAPWKKIQQNTFTRWCNEHLKCV
SKRIANLQTDLSDGLRLIALLEVLSQKKMHRKHNQRPTFRQMQLENVSVALEFLDRESIK LVSIDSKAIVDGNLKLILGLIWTLILHYSISMPMWDEEEDEEAKKQTPKQRLLGWIQNKL PQLPITNFSRDWQSGRALGALVDSCAPGLCPDWDSWDASKPVTNAREAMQQADDWLGIPQ VITPEEIVDPNVDEHSVMTYLSQFPKAKLKPGAPLRPKLNPKKARAYGPGIEPTGNMVKK RAEFTVETRSAGQGEVLVYVEDPAGHQEEAKVTANNDKNRTFSVWYVPEVTGTHKVTVLF AGQHIAKSPFEVYVDKSQGDASKVTAQGPGLEPSGNIANKTTYFEIFTAGAGTGEVEVVI QDPMGQKGTVEPQLEARGDSTYRCSYQPTMEGVHTVHVTFAGVPIPRSPYTVTVGQACNP SACRAVGRGLQPKGVRVKETADFKVYTKGAGSGELKVTVKGPKGEERVKQKDLGDGVYGF EYYPMVPGTYIVTITWGGQNIGRSPFEVKVGTECGNQKVRAWGPGLEGGVVGKSADFVVE AIGDDVGTLGFSVEGPSQAKIECDDKGDGSCDVRYWPQEAGEYAVHVLCNSEDIRLSPFM ADIRDAPQDFHPDRVKARGPGLEKTGVAVNKPAEFTVDAKHGGKAPLRVQVQDNEGCPVE ALVKDNGNGTYSCSYVPRKPVKHTAMVSWGGVSIPNSPFRVNVGAGSHPNKVKVYGPGVA KTGLKAHEPTYFTVDCAEAGQGDVSIGIKCAPGVVGPAEADIDFDIIRNDNDTFTVKYTP RGAGSYTIMVLFADQATPTSPIRVKVEPSHDASKVKAEGPGLSRTGVELGKPTHFTVNAK AAGKGKLDVQFSGLTKGDAVRDVDIIDHHDNTYTVKYTPVQQGPVGVNVTYGGDPIPKSP FSVAVSPSLDLSKIKVSGLGEKVDVGKDQEFTVKSKGAGGQGKVASKIVGPSGAAVPCKV EPGLGADNSVVRFLPREEGPYEVEVTYDGVPVPGSPFPLEAVAPTKPSKVKAFGPGLQGG SAGSPARFTIDTKGAGTGGLGLTVEGPCEAQLECLDNGDGTCSVSYVPTEPGDYNINILF ADTHIPGSPFKAHVVPCFDASKVKCSGPGLERATAGEVGQFQVDCSSAGSAELTIEICSE AGLPAEVYIQDHGDGTHTITYIPLCPGAYTVTIKYGGQPVPNFPSKLQVEPAVDTSGVQC YGPGIEGQGVFREATTEFSVDARALTQTGGPHVKARVANPSGNLTETYVQDRGDGMYKVE YTPYEEGLHSVDVTYDGSPVPSSPFQVPVTEGCDPSRVRVHGPGIQSGTTNKPNKFTVET RGAGTGGLGLAVEGPSEAKMSCMDNKDGSCSVEYIPYEAGTYSLNVTYGGHQVPGSPFKV PVHDVTDASKVKCSGPGLSPGMVRANLPQSFQVDTSKAGVAPLQVKVQGPKGLVEPVDVV DNADGTQTVNYVPSREGPYSISVLYGDEEVPRSPFKVKVLPTHDASKVKASGPGLNTTGV PASLPVEFTIDAKDAGEGLLAVQITDPEGKPKKTHIQDNHDGTYTVAYVPDVTGRYTILI KYGGDEIPFSPYRVRAVPTGDASKCTVTVSIGGHGLGAGIGPTIQIGEETVITVDTKAAG KGKVTCTVCTPDGSEVDVDVVENEDGTFDIFYTAPQPGKYVICVRFGGEHVPNSPFQVTA LAGDQPSVQPPLRSQQLAPQYTYAQGGQQTWAPERPLVGVNGLDVTSLRPFDLVIPFTIK KGEITGEVRMPSGKVAQPTITDNKDGTVTVRYAPSEAGLHEMDIRYDNMHIPGSPLQFYV DYVNCGHVTAYGPGLTHGVVNKPATFTVNTKDAGEGGLSLAIEGPSKAEISCTDNQDGTC SVSYLPVLPGDYSILVKYNEQHVPGSPFTARVTGDDSMRMSHLKVGSAADIPINISETDL SLLTATVVPPSGREEPCLLKRLRNGHVGISFVPKETGEHLVHVKKNGQHVASSPIPVVIS QSEIGDASRVRVSGQGLHEGHTFEPAEFIIDTRDAGYGGLSLSIEGPSKVDINTEDLEDG TCRVTYCPTEPGNYIINIKFADQHVPGSPFSVKVTGEGRVKESITRRRRAPSVANVGSHC DLSLKIPEISIQDMTAQVTSPSGKTHEAEIVEGENHTYCIRFVPAEMGTHTVSVKYKGQH VPGSPFQFTVGPLGEGGAHKVRAGGPGLERAEAGVPAEFSIWTREAGAGGLAIAVEGPSK AEISFEDRKDGSCGVAYVVQEPGDYEVSVKFNEEHIPDSPFVVPVASPSGDARRLTVSSL QESGLKVNQPASFAVSLNGAKGAIDAKVHSPSGALEECYVTEIDQDKYAVRFIPRENGVY LIDVKFNGTHIPGSPFKIRVGEPGHGGDPGLVSAYGAGLEGGVTGNPAEFVVNTSNAGAG ALSVTIDGPSKVKMDCQECPEGYRVTYTPMAPGSYLISIKYGGPYHIGGSPFKAKVTGPR LVSNHSLHETSSVFVDSLTKATCAPQHGAPGPGPADASKVVAKGLGLSKAYVGQKSSFTV DCSKAGNNMLLVGVHGPRTPCEEILVKHVGSRLYSVSYLLKDKGEYTLVVKWGDEHIPGS PYRVVVP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Glutathione | Ligand Info | |||||
| Structure Description | Crystal structure of the filamin A repeat 21 complexed with the integrin beta7 cytoplasmic tail peptide | PDB:2BRQ | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | No | [3] |
| PDB Sequence |
GGAHKVRAGG
2245 PGLERAEAGV2255 PAEFSIWTRE2265 AGAGGLAIAV2275 EGPSKAEISF2285 EDRKDGSCGV 2295 AYVVQEPGDY2305 EVSVKFNEEH2315 IPDSPFVVPV2325 ASPS
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |
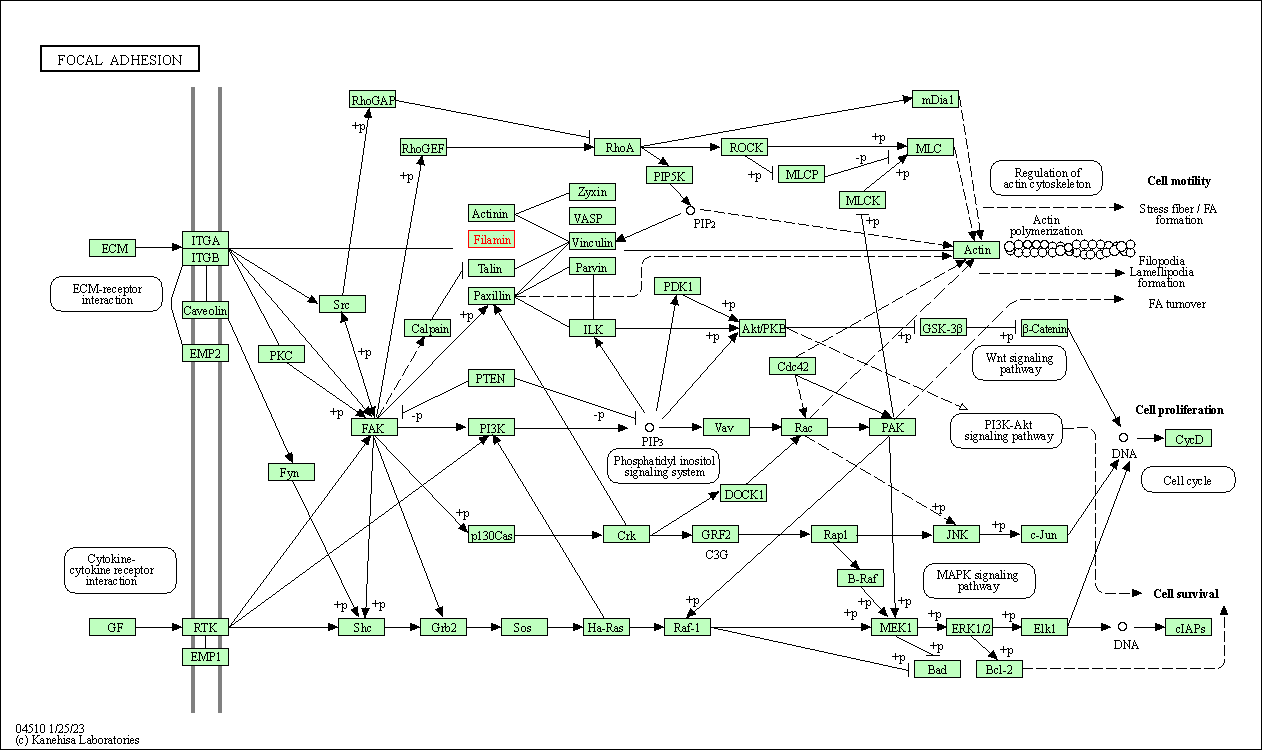
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Degree | 20 | Degree centrality | 2.15E-03 | Betweenness centrality | 1.33E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.34E-01 | Radiality | 1.41E+01 | Clustering coefficient | 4.74E-02 |
| Neighborhood connectivity | 2.28E+01 | Topological coefficient | 7.00E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT04388254) Simufilam (PTI-125), 100 mg, for Mild-to-moderate Alzheimer's Disease Patients (PTI-125). U.S. National Institutes of Health. | |||||
| REF 3 | The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006 Feb 3;21(3):337-47. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

