Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T70490
(Former ID: TTDR00571)
|
|||||
| Target Name |
Dipeptidyl peptidase I (CTSC)
|
|||||
| Synonyms |
Dipeptidyl transferase; Dipeptidyl peptidase 1; DPPI; DPP-I; Cysteine protease dipeptidyl peptidase I; Cathepsin J; Cathepsin C; CPPI
Click to Show/Hide
|
|||||
| Gene Name |
CTSC
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Bronchiectasis [ICD-11: CA24] | |||||
| Function |
Has dipeptidylpeptidase activity. Active against a broad range of dipeptide substrates composed of both polar and hydrophobic amino acids. Proline cannot occupy the P1 position and arginine cannot occupy the P2 position of the substrate. Can act as both an exopeptidase and endopeptidase. Activates serine proteases such as elastase, cathepsin G and granzymes A and B. Can also activate neuraminidase and factor XIII. Thiol protease.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.14.1
|
|||||
| Sequence |
MGAGPSLLLAALLLLLSGDGAVRCDTPANCTYLDLLGTWVFQVGSSGSQRDVNCSVMGPQ
EKKVVVYLQKLDTAYDDLGNSGHFTIIYNQGFEIVLNDYKWFAFFKYKEEGSKVTTYCNE TMTGWVHDVLGRNWACFTGKKVGTASENVYVNIAHLKNSQEKYSNRLYKYDHNFVKAINA IQKSWTATTYMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWRNV HGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQTPILSPQEVVSCSQYAQGCEG GFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEALM KLELVHHGPMAVAFEVYDDFLHYKKGIYHHTGLRDPFNPFELTNHAVLLVGYGTDSASGM DYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATPIPKL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T93R42 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-tyrosine | Ligand Info | |||||
| Structure Description | Crystal structure of Cathepsin C in complex with dipeptide substrates | PDB:4OEL | ||||
| Method | X-ray diffraction | Resolution | 1.40 Å | Mutation | Yes | [5] |
| PDB Sequence |
> Chain A
DTPANCTYLD 10 LLGTWVFQVG20 SSGSQRDVNC30 SVMGPQEKKV40 VVYLQKLDTA50 YDDLGNSGHF 60 TIIYNQGFEI70 VLNDYKWFAF80 FKYKEEGSKV90 TTYCNETMTG100 WVHDVLGRNW 110 ACFTGKKVLH206 LPTSWDWRNV216 HGINFVSPVR226 NQASCGSCYS236 FASMGMLEAR 246 IRILTNNSQT256 PILSPQEVVS266 CSQYAQGCEG276 GFPYLIAGKY286 AQDFGLVEEA 296 CFPYTGTDSP306 CKMKEDCFRY316 YSSEYHYVGG326 FYGGCNEALM336 KLELVHHGPM 346 AVAFEVYDDF356 LHYKKGIYHH366 T> Chain B PFNPFELTNH 381 AVLLVGYGTD391 SASGMDYWIV401 KNSWGTGWGE411 NGYFRIRRGT421 DECAIESIAV 431 AATPIPKLE
|
|||||
|
|
||||||
| Ligand Name: L-serine | Ligand Info | |||||
| Structure Description | Crystal structure of Cathepsin C in complex with dipeptide substrates | PDB:4OEL | ||||
| Method | X-ray diffraction | Resolution | 1.40 Å | Mutation | Yes | [5] |
| PDB Sequence |
> Chain A
DTPANCTYLD 10 LLGTWVFQVG20 SSGSQRDVNC30 SVMGPQEKKV40 VVYLQKLDTA50 YDDLGNSGHF 60 TIIYNQGFEI70 VLNDYKWFAF80 FKYKEEGSKV90 TTYCNETMTG100 WVHDVLGRNW 110 ACFTGKKVLH206 LPTSWDWRNV216 HGINFVSPVR226 NQASCGSCYS236 FASMGMLEAR 246 IRILTNNSQT256 PILSPQEVVS266 CSQYAQGCEG276 GFPYLIAGKY286 AQDFGLVEEA 296 CFPYTGTDSP306 CKMKEDCFRY316 YSSEYHYVGG326 FYGGCNEALM336 KLELVHHGPM 346 AVAFEVYDDF356 LHYKKGIYHH366 T> Chain B PFNPFELTNH 381 AVLLVGYGTD391 SASGMDYWIV401 KNSWGTGWGE411 NGYFRIRRGT421 DECAIESIAV 431 AATPIPKLE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Lysosome | hsa04142 | Affiliated Target |
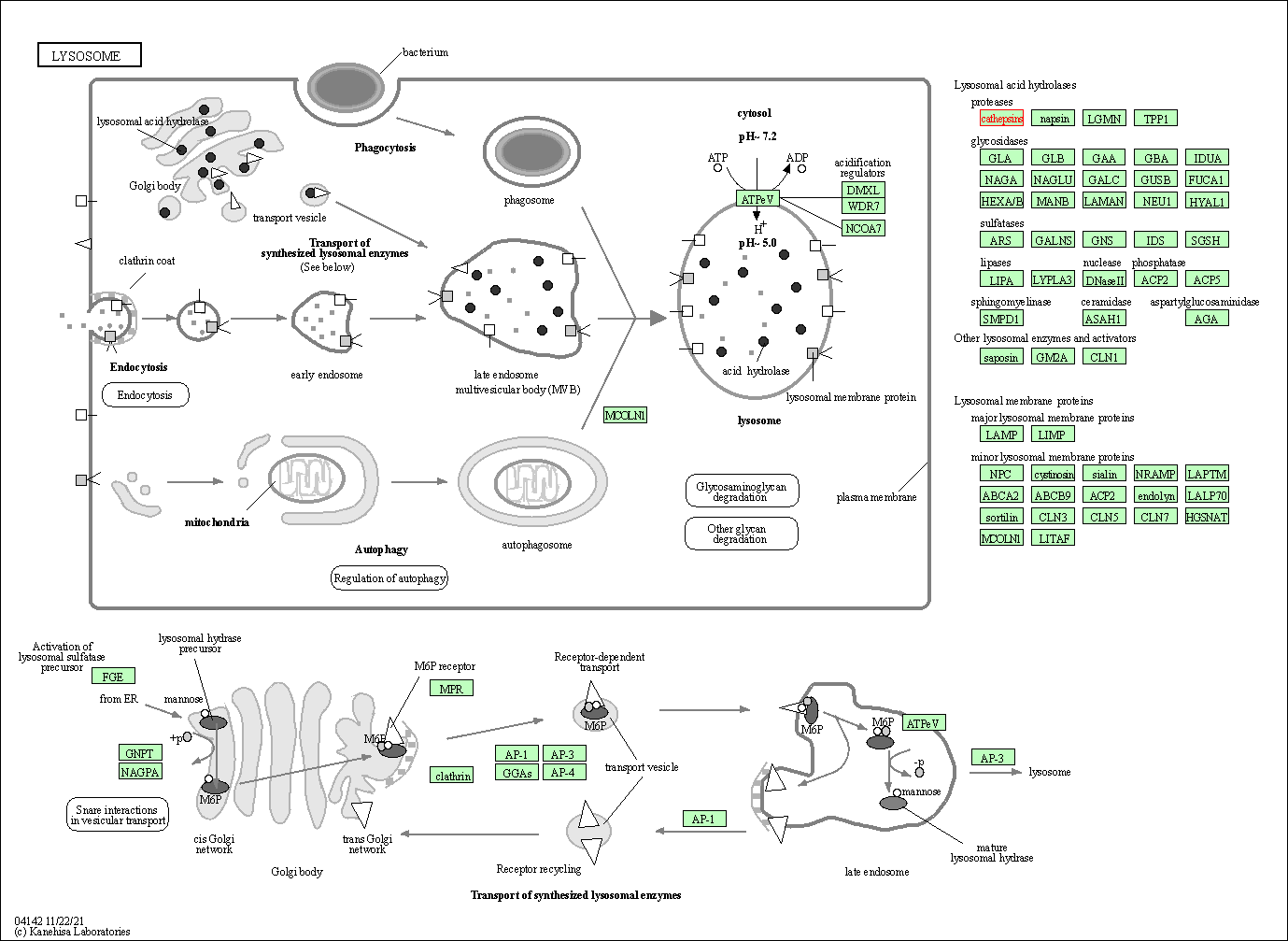
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
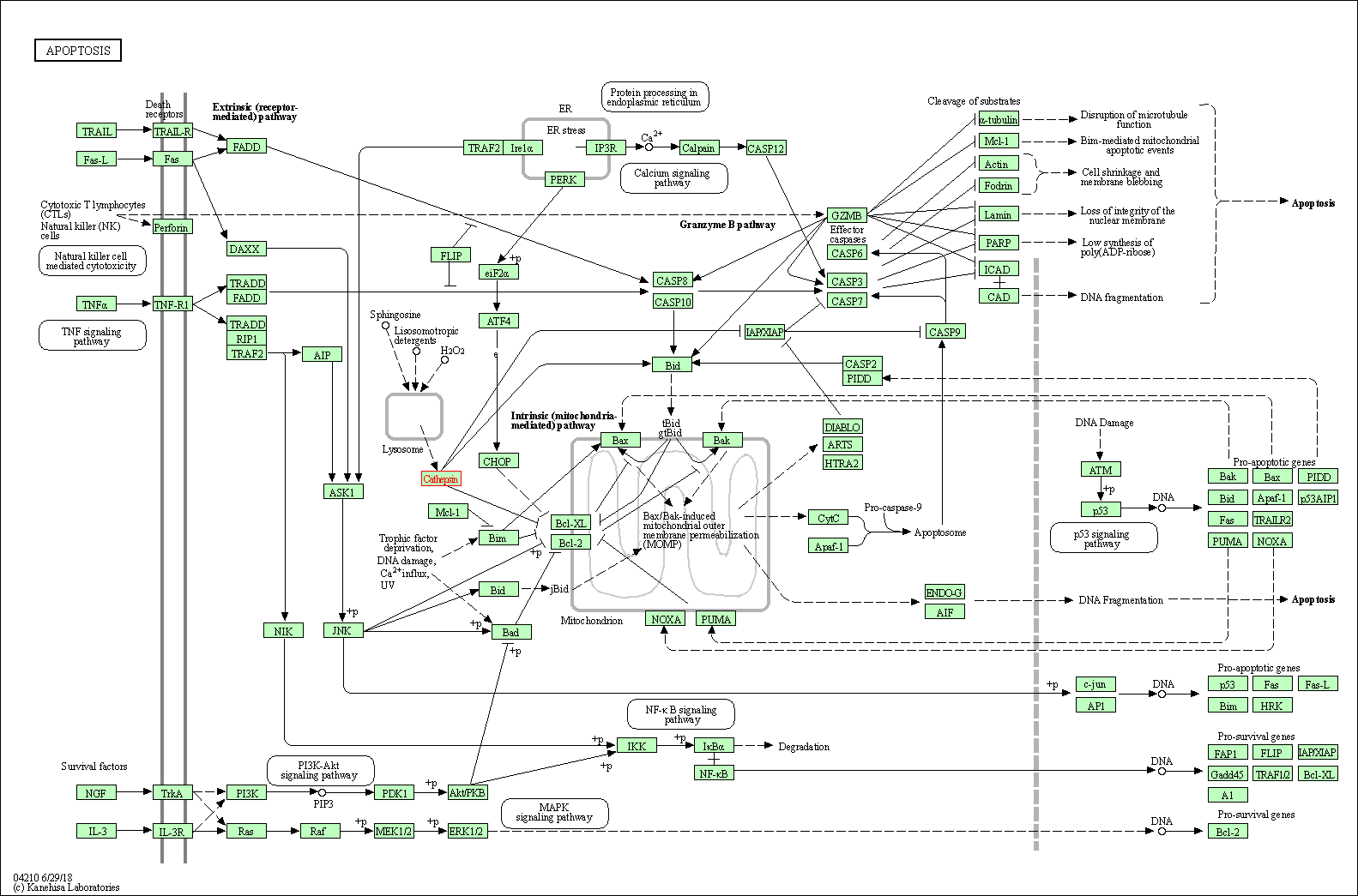
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Lysosome | |||||
| NetPath Pathway | [+] 3 NetPath Pathways | + | ||||
| 1 | IL2 Signaling Pathway | |||||
| 2 | IL4 Signaling Pathway | |||||
| 3 | TGF_beta_Receptor Signaling Pathway | |||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | COPII (Coat Protein 2) Mediated Vesicle Transport | |||||
| 2 | MHC class II antigen presentation | |||||
| 3 | Cargo concentration in the ER | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | miR-targeted genes in muscle cell - TarBase | |||||
| 2 | miR-targeted genes in lymphocytes - TarBase | |||||
| 3 | miR-targeted genes in leukocytes - TarBase | |||||
| 4 | miR-targeted genes in epithelium - TarBase | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ClinicalTrials.gov (NCT02058407) A Study to Evaluate the Safety, Tolerability, Pharmacokinetics (PK), Pharmacodynamics (PD) and Food Effect of Single or Repeat Doses of GSK2793660 in Healthy Subjects. U.S. National Institutes of Health. | |||||
| REF 2 | ClinicalTrials.gov (NCT05238675) A Randomised, Double-blind, Placebo-controlled, Parallel Group, Dose-finding Study Evaluating Efficacy, Safety and Tolerability of BI 1291583 qd Over at Least 24 Weeks in Patients With Bronchiectasis (AirleafTM). U.S.National Institutes of Health. | |||||
| REF 3 | Substrate optimization for monitoring cathepsin C activity in live cells. Bioorg Med Chem. 2009 Feb 1;17(3):1064-70. | |||||
| REF 4 | Carboxyl-modified amino acids and peptides as protease inhibitors. J Med Chem. 1986 Jan;29(1):104-11. | |||||
| REF 5 | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis. Biochemistry. 2012 Sep 25;51(38):7551-68. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

