Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T76910
(Former ID: TTDS00136)
|
|||||
| Target Name |
Glycogen synthase kinase-3 alpha (GSK-3A)
|
|||||
| Synonyms |
Serine/threonineprotein kinase GSK3A; Serine/threonine-protein kinase GSK3A; Glycogen synthase kinase3 alpha; Glycogen synthase kinase 3; GSK3 alpha; GSK-3 alpha; GSK-3
Click to Show/Hide
|
|||||
| Gene Name |
GSK3A
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Epilepsy/seizure [ICD-11: 8A61-8A6Z] | |||||
| Function |
Requires primed phosphorylation of the majority of its substrates. Contributes to insulin regulation of glycogen synthesis by phosphorylating and inhibiting GYS1 activity and hence glycogen synthesis. Regulates glycogen metabolism in liver, but not in muscle. May also mediate the development of insulin resistance by regulating activation of transcription factors. In Wnt signaling, regulates the level and transcriptional activity of nuclear CTNNB1/beta-catenin. Facilitates amyloid precursor protein (APP) processing and the generation of APP-derived amyloid plaques found in Alzheimer disease. May be involved in the regulation of replication in pancreatic beta-cells. Is necessary for the establishment of neuronal polarity and axon outgrowth. Through phosphorylation of the anti-apoptotic protein MCL1, may control cell apoptosis in response to growth factors deprivation. Acts as a regulator of autophagy by mediating phosphorylation of KAT5/TIP60 under starvation conditions, leading to activate KAT5/TIP60 acetyltransferase activity and promote acetylation of key autophagy regulators, such as ULK1 and RUBCNL/Pacer. Constitutively active protein kinase that acts as a negative regulator in the hormonal control of glucose homeostasis, Wnt signaling and regulation of transcription factors and microtubules, by phosphorylating and inactivating glycogen synthase (GYS1 or GYS2), CTNNB1/beta-catenin, APC and AXIN1.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.26
|
|||||
| Sequence |
MSGGGPSGGGPGGSGRARTSSFAEPGGGGGGGGGGPGGSASGPGGTGGGKASVGAMGGGV
GASSSGGGPGGSGGGGSGGPGAGTSFPPPGVKLGRDSGKVTTVVATLGQGPERSQEVAYT DIKVIGNGSFGVVYQARLAETRELVAIKKVLQDKRFKNRELQIMRKLDHCNIVRLRYFFY SSGEKKDELYLNLVLEYVPETVYRVARHFTKAKLTIPILYVKVYMYQLFRSLAYIHSQGV CHRDIKPQNLLVDPDTAVLKLCDFGSAKQLVRGEPNVSYICSRYYRAPELIFGATDYTSS IDVWSAGCVLAELLLGQPIFPGDSGVDQLVEIIKVLGTPTREQIREMNPNYTEFKFPQIK AHPWTKVFKSRTPPEAIALCSSLLEYTPSSRLSPLEACAHSFFDELRCLGTQLPNNRPLP PLFNFSAGELSIQPSLNAILIPPHLRSPAGTTTLTPSSQALTETPTSSDWQSTDATPTLT NSS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Patented Agent(s) | [+] 3 Patented Agents | + | ||||
| 1 | AR-A014418 | Drug Info | Patented | Ovarian cancer | [8] | |
| 2 | PMID27828716-Compound-BIO-acetoxime | Drug Info | Patented | Malignant glioma | [8] | |
| 3 | TDZD-8 | Drug Info | Patented | Malignant glioma | [8] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 27 Inhibitor drugs | + | ||||
| 1 | Indirubin derivative 2 | Drug Info | [8] | |||
| 2 | KENPAULLONE | Drug Info | [8] | |||
| 3 | Maleimides derivative 1 | Drug Info | [8] | |||
| 4 | Maleimides derivative 3 | Drug Info | [8] | |||
| 5 | PMID27828716-Compound-16 | Drug Info | [8] | |||
| 6 | PMID27828716-Compound-17 | Drug Info | [8] | |||
| 7 | PMID27828716-Compound-18 | Drug Info | [8] | |||
| 8 | PMID27828716-Compound-19 | Drug Info | [8] | |||
| 9 | PMID27828716-Compound-20 | Drug Info | [8] | |||
| 10 | PMID27828716-Compound-21 | Drug Info | [8] | |||
| 11 | PMID27828716-Compound-BIO-acetoxime | Drug Info | [8] | |||
| 12 | Pyrazole and benzimidazole derivative 1 | Drug Info | [8] | |||
| 13 | Pyrazolodihydropyridine derivative 1 | Drug Info | [8] | |||
| 14 | Quinoline derivative 14 | Drug Info | [8] | |||
| 15 | Quinoline derivative 15 | Drug Info | [8] | |||
| 16 | Quinolinyl pyrazinyl urea derivative 1 | Drug Info | [8] | |||
| 17 | Quinolinyl pyrazinyl urea derivative 2 | Drug Info | [8] | |||
| 18 | Spiroquinolone derivative 1 | Drug Info | [8] | |||
| 19 | TDZD-8 | Drug Info | [8] | |||
| 20 | Thiadiazolidindione derivative 1 | Drug Info | [8] | |||
| 21 | Thiadiazolidindione derivative 2 | Drug Info | [8] | |||
| 22 | Thiadiazolidindione derivative 3 | Drug Info | [8] | |||
| 23 | 6-bromoindirubin-3-oxime | Drug Info | [9] | |||
| 24 | AR-534 | Drug Info | [10] | |||
| 25 | Benzofuran-3-yl-(indol-3-yl)maleimides | Drug Info | [11] | |||
| 26 | Lithium chloride | Drug Info | [12], [13] | |||
| 27 | SB216763 | Drug Info | [14], [15], [16] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Chemokine signaling pathway | hsa04062 | Affiliated Target |
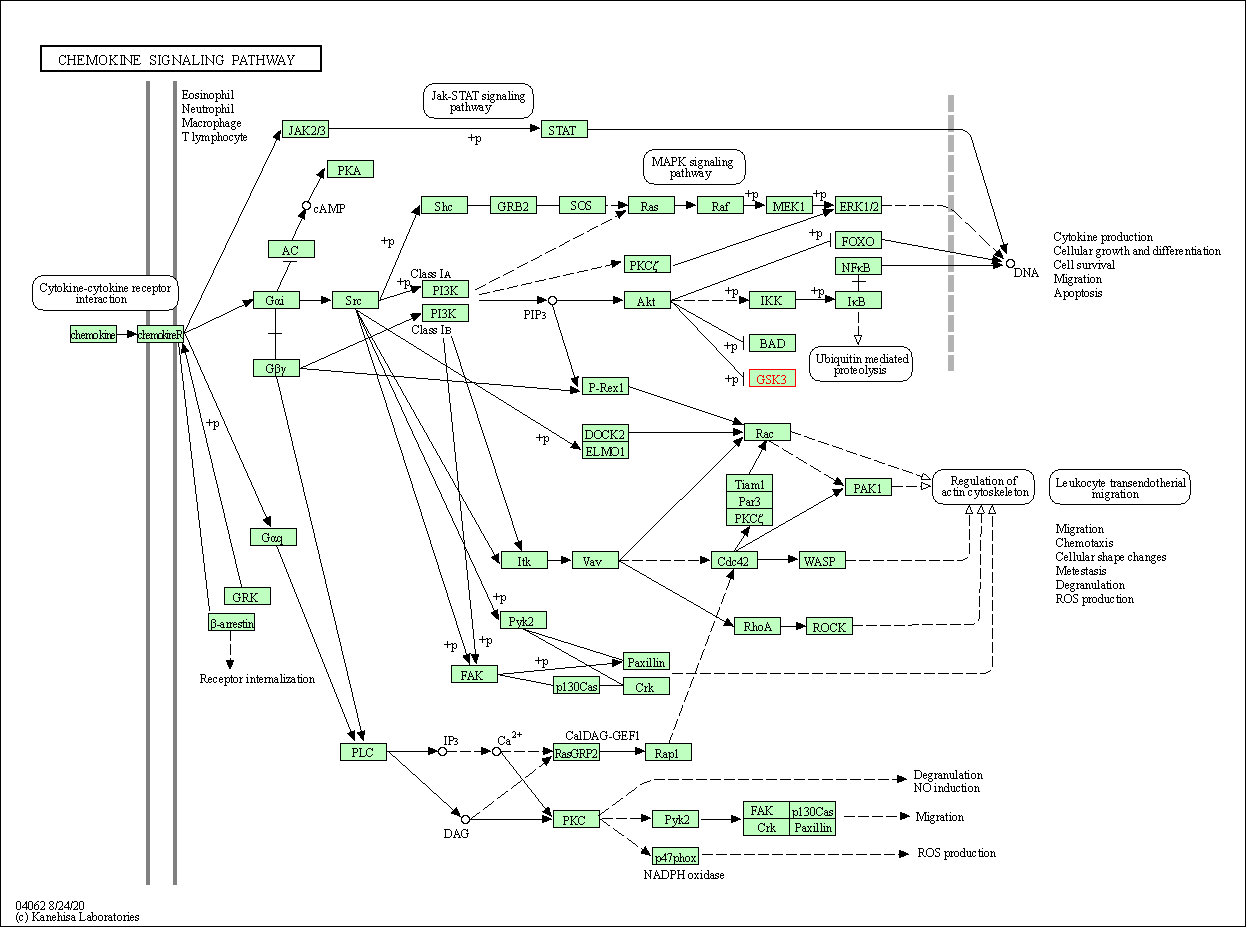
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
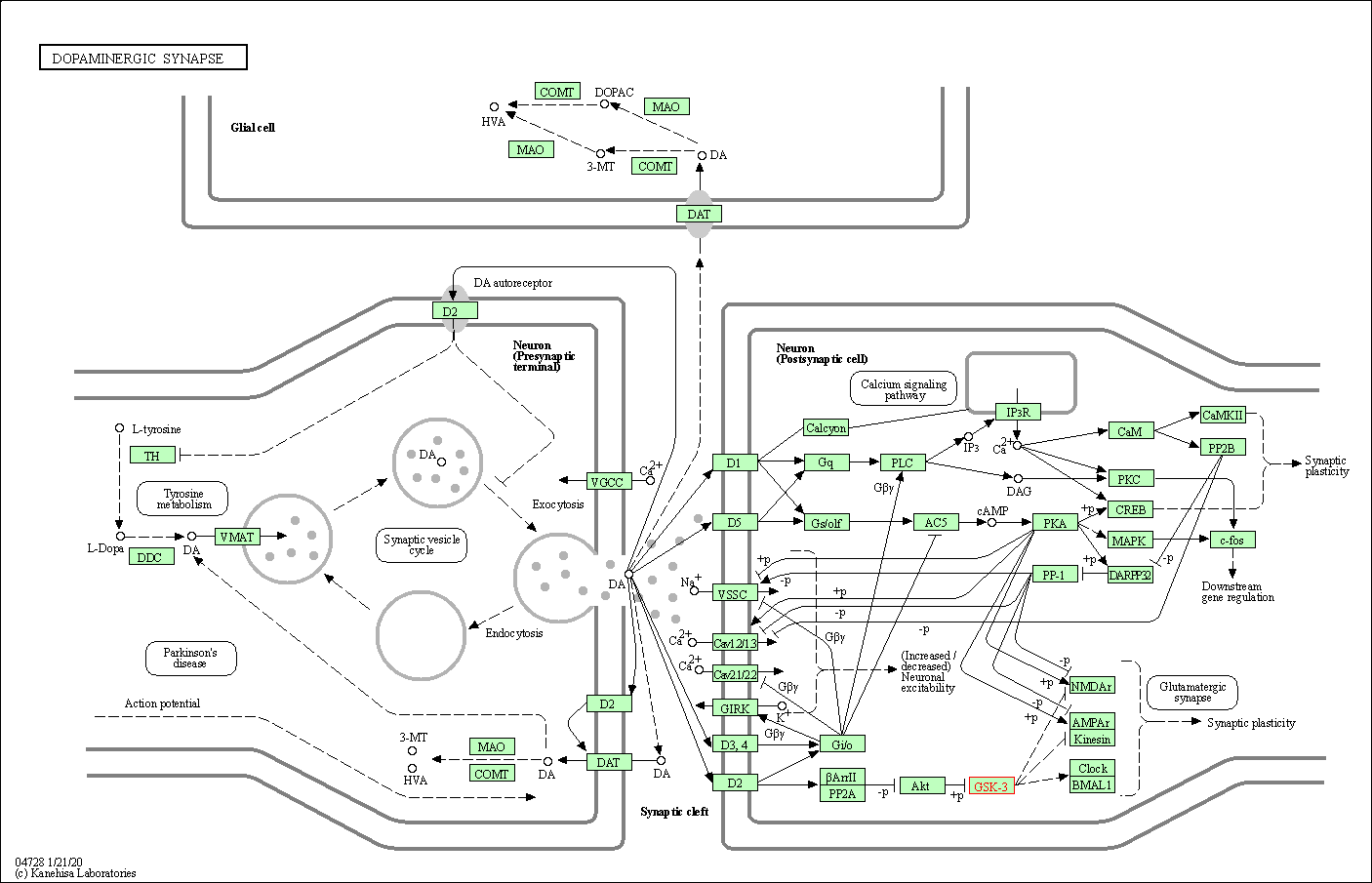
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 5.64E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.41E-01 | Radiality | 1.42E+01 | Clustering coefficient | 4.29E-01 |
| Neighborhood connectivity | 6.86E+01 | Topological coefficient | 1.65E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | TGF_beta_Receptor Signaling Pathway | |||||
| Panther Pathway | [+] 4 Panther Pathways | + | ||||
| 1 | Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | |||||
| 2 | Insulin/IGF pathway-protein kinase B signaling cascade | |||||
| 3 | PDGF signaling pathway | |||||
| 4 | Ras Pathway | |||||
| PID Pathway | [+] 4 PID Pathways | + | ||||
| 1 | Degradation of beta catenin | |||||
| 2 | Canonical Wnt signaling pathway | |||||
| 3 | FOXM1 transcription factor network | |||||
| 4 | Class I PI3K signaling events mediated by Akt | |||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | AKT phosphorylates targets in the cytosol | |||||
| 2 | XBP1(S) activates chaperone genes | |||||
| 3 | Constitutive Signaling by AKT1 E17K in Cancer | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Challenges and new opportunities in the investigation of new drug therapies to treat frontotemporal dementia. Expert Opin Ther Targets. 2008 Nov;12(11):1367-76. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7009). | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | Emerging disease-modifying therapies for the treatment of motor neuron disease/amyotropic lateral sclerosis. Expert Opin Emerg Drugs. 2007 May;12(2):229-52. | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7958). | |||||
| REF 6 | ClinicalTrials.gov (NCT01214603) A Study in Participants With Acute Leukemia. U.S. National Institutes of Health. | |||||
| REF 7 | Open-label treatment trial of lithium to target the underlying defect in fragile X syndrome. J Dev Behav Pediatr. 2008 Aug;29(4):293-302. | |||||
| REF 8 | Glycogen synthase kinase 3 (GSK-3) inhibitors: a patent update (2014-2015).Expert Opin Ther Pat. 2017 Jun;27(6):657-666. | |||||
| REF 9 | Stable generation of serum- and feeder-free embryonic stem cell-derived mice with full germline-competency by using a GSK3 specific inhibitor. Genesis. 2009 Jun;47(6):414-22. | |||||
| REF 10 | A zebrafish model of tauopathy allows in vivo imaging of neuronal cell death and drug evaluation. J Clin Invest. 2009 May;119(5):1382-95. | |||||
| REF 11 | From a natural product lead to the identification of potent and selective benzofuran-3-yl-(indol-3-yl)maleimides as glycogen synthase kinase 3beta inhibitors that suppress proliferation and survival of pancreatic cancer cells. J Med Chem. 2009 Apr 9;52(7):1853-63. | |||||
| REF 12 | Glycogen synthase kinase 3 beta (GSK-3 beta) as a therapeutic target in neuroAIDS. J Neuroimmune Pharmacol. 2007 Mar;2(1):93-6. | |||||
| REF 13 | Glycogen synthase kinase 3: an emerging therapeutic target. Trends Mol Med. 2002 Mar;8(3):126-32. | |||||
| REF 14 | Intracellular protein phosphorylation in eosinophils and the functional relevance in cytokine production. Int Arch Allergy Immunol. 2009;149 Suppl 1:45-50. | |||||
| REF 15 | Cocaine-induced hyperactivity and sensitization are dependent on GSK3. Neuropharmacology. 2009 Jun;56(8):1116-23. | |||||
| REF 16 | The ceiling effect of pharmacological postconditioning with the phytoestrogen genistein is reversed by the GSK3beta inhibitor SB 216763 [3-(2,4-dic... J Pharmacol Exp Ther. 2009 Jun;329(3):1134-41. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

