Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T89055
(Former ID: TTDNC00613)
|
|||||
| Target Name |
ERK activator kinase 2 (MEK2)
|
|||||
| Synonyms |
PRKMK2; MKK2; MEK 2; MAPKK 2; MAPK/ERK kinase 2; MAP kinase kinase 2; Dual specificity mitogenactivated protein kinase kinase 2; Dual specificity mitogen-activated protein kinase kinase 2
Click to Show/Hide
|
|||||
| Gene Name |
MAP2K2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Melanoma [ICD-11: 2C30] | |||||
| 2 | Thyroid cancer [ICD-11: 2D10] | |||||
| Function |
Activates the ERK1 and ERK2 MAP kinases. Catalyzes the concomitant phosphorylation of a threonine and a tyrosine residue in a Thr-Glu-Tyr sequence located in MAP kinases.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.12.2
|
|||||
| Sequence |
MLARRKPVLPALTINPTIAEGPSPTSEGASEANLVDLQKKLEELELDEQQKKRLEAFLTQ
KAKVGELKDDDFERISELGAGNGGVVTKVQHRPSGLIMARKLIHLEIKPAIRNQIIRELQ VLHECNSPYIVGFYGAFYSDGEISICMEHMDGGSLDQVLKEAKRIPEEILGKVSIAVLRG LAYLREKHQIMHRDVKPSNILVNSRGEIKLCDFGVSGQLIDSMANSFVGTRSYMAPERLQ GTHYSVQSDIWSMGLSLVELAVGRYPIPPPDAKELEAIFGRPVVDGEEGEPHSISPRPRP PGRPVSGHGMDSRPAMAIFELLDYIVNEPPPKLPNGVFTPDFQEFVNKCLIKNPAERADL KMLTNHTFIKRSEVEEVDFAGWLCKTLRLNQPGTPTRTAV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T66S13 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | RDEA-436 | Drug Info | Discontinued in Phase 1 | Solid tumour/cancer | [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | RDEA-436 | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | X-ray structure of the human mitogen-activated protein kinase kinase 2 (MEK2)in a complex with ligand and MgATP | PDB:1S9I | ||||
| Method | X-ray diffraction | Resolution | 3.20 Å | Mutation | No | [5] |
| PDB Sequence |
QKAKVGELKD
69 DDFERISELG79 AGNGGVVTKV89 QHRPSGLIMA99 RKLIHLEIKP109 AIRNQIIREL 119 QVLHECNSPY129 IVGFYGAFYS139 DGEISICMEH149 MDGGSLDQVL159 KEAKRIPEEI 169 LGKVSIAVLR179 GLAYLREKHQ189 IMHRDVKPSN199 ILVNSRGEIK209 LCDFGVSGQL 219 IDSMVGTRSY233 MAPERLQGTH243 YSVQSDIWSM253 GLSLVELAVG263 RYPIPPPDAK 273 ELEAIFGRPV283 VDRPAMAIFE320 LLDYIVNEPP330 PKLPNGVFTP340 DFQEFVNKCL 350 IKNPAERADL360 KMLTNHTFIK370 RSEVEEVDFA380 GWLCKTLRLN390 QPG |
|||||
|
|
LEU78
3.604
GLY79
3.766
ALA80
3.733
GLY81
3.294
ASN82
3.329
GLY84
3.705
VAL85
4.565
VAL86
3.315
ALA99
3.651
LYS101
2.590
VAL131
4.165
MET147
3.368
GLU148
2.913
|
|||||
| Ligand Name: 5-{3,4-Difluoro-2-[(2-fluoro-4-iodophenyl)amino]phenyl}-N-(2-morpholin-4-ylethyl)-1,3,4-oxadiazol-2-amine | Ligand Info | |||||
| Structure Description | X-ray structure of the human mitogen-activated protein kinase kinase 2 (MEK2)in a complex with ligand and MgATP | PDB:1S9I | ||||
| Method | X-ray diffraction | Resolution | 3.20 Å | Mutation | No | [5] |
| PDB Sequence |
QKAKVGELKD
69 DDFERISELG79 AGNGGVVTKV89 QHRPSGLIMA99 RKLIHLEIKP109 AIRNQIIREL 119 QVLHECNSPY129 IVGFYGAFYS139 DGEISICMEH149 MDGGSLDQVL159 KEAKRIPEEI 169 LGKVSIAVLR179 GLAYLREKHQ189 IMHRDVKPSN199 ILVNSRGEIK209 LCDFGVSGQL 219 IDSMVGTRSY233 MAPERLQGTH243 YSVQSDIWSM253 GLSLVELAVG263 RYPIPPPDAK 273 ELEAIFGRPV283 VDRPAMAIFE320 LLDYIVNEPP330 PKLPNGVFTP340 DFQEFVNKCL 350 IKNPAERADL360 KMLTNHTFIK370 RSEVEEVDFA380 GWLCKTLRLN390 QPG |
|||||
|
|
GLY81
4.747
ASN82
3.131
GLY83
4.692
LYS101
3.063
ILE103
4.917
LEU119
3.258
LEU122
3.620
ILE130
4.871
VAL131
3.250
GLY132
3.953
PHE133
4.781
ILE145
3.745
MET147
3.479
ARG193
3.540
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| ErbB signaling pathway | hsa04012 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |
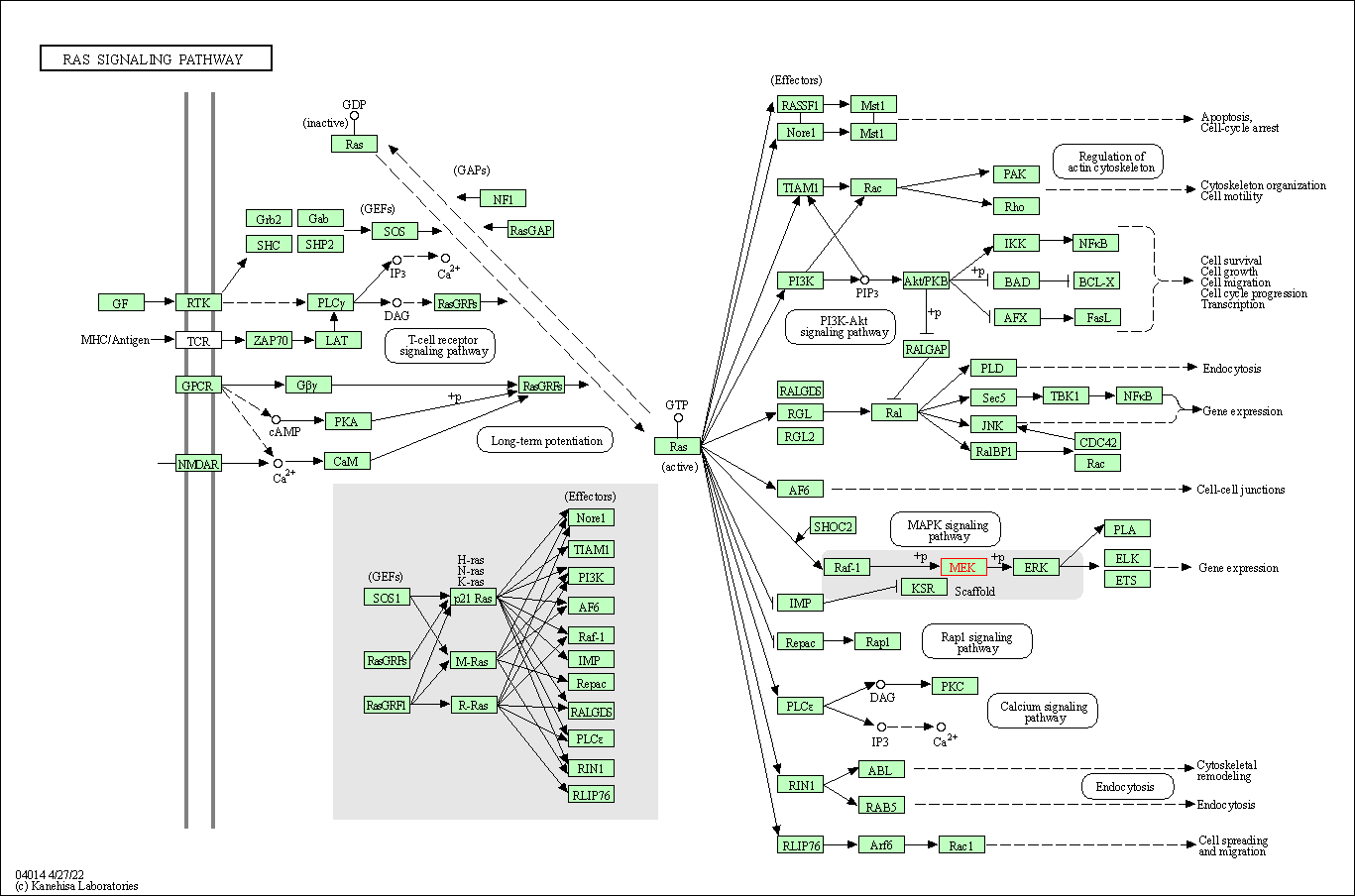
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |
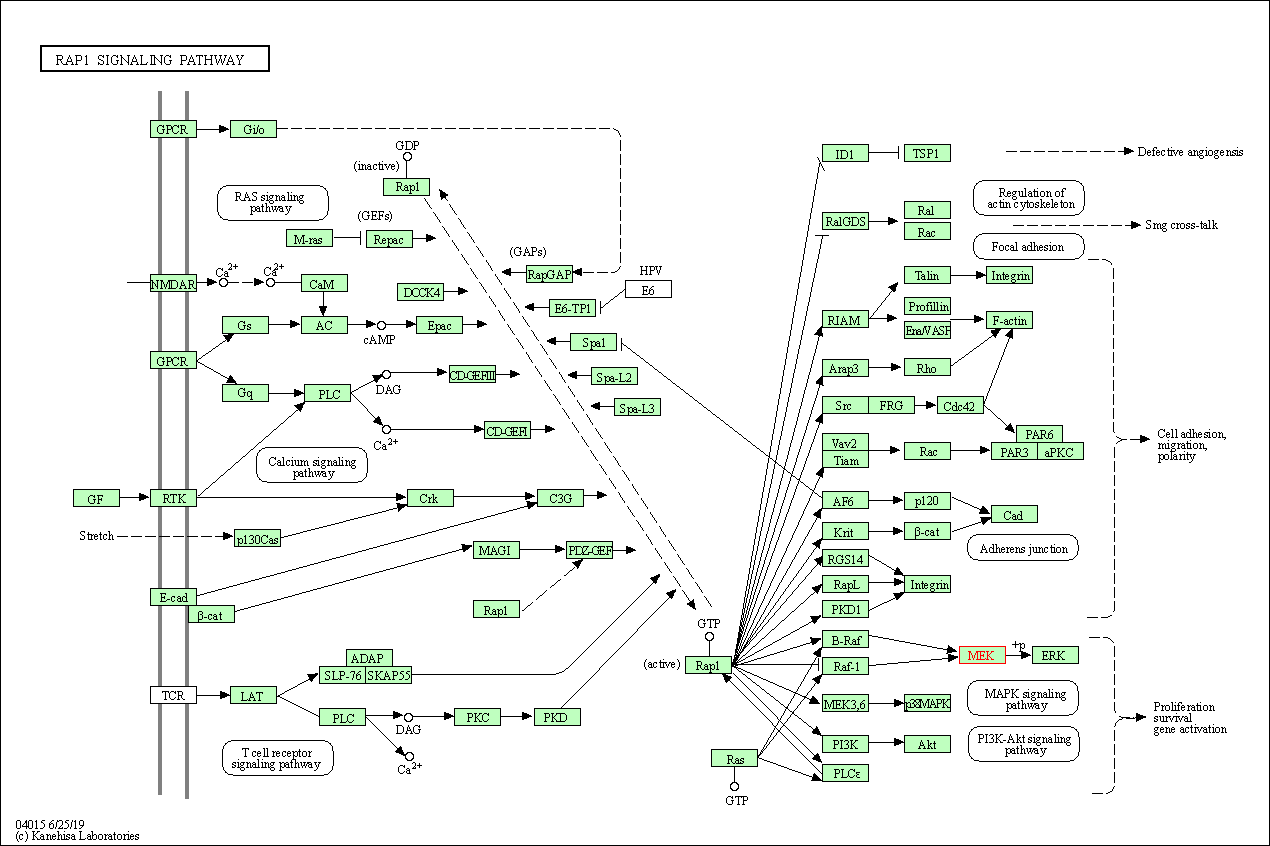
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |
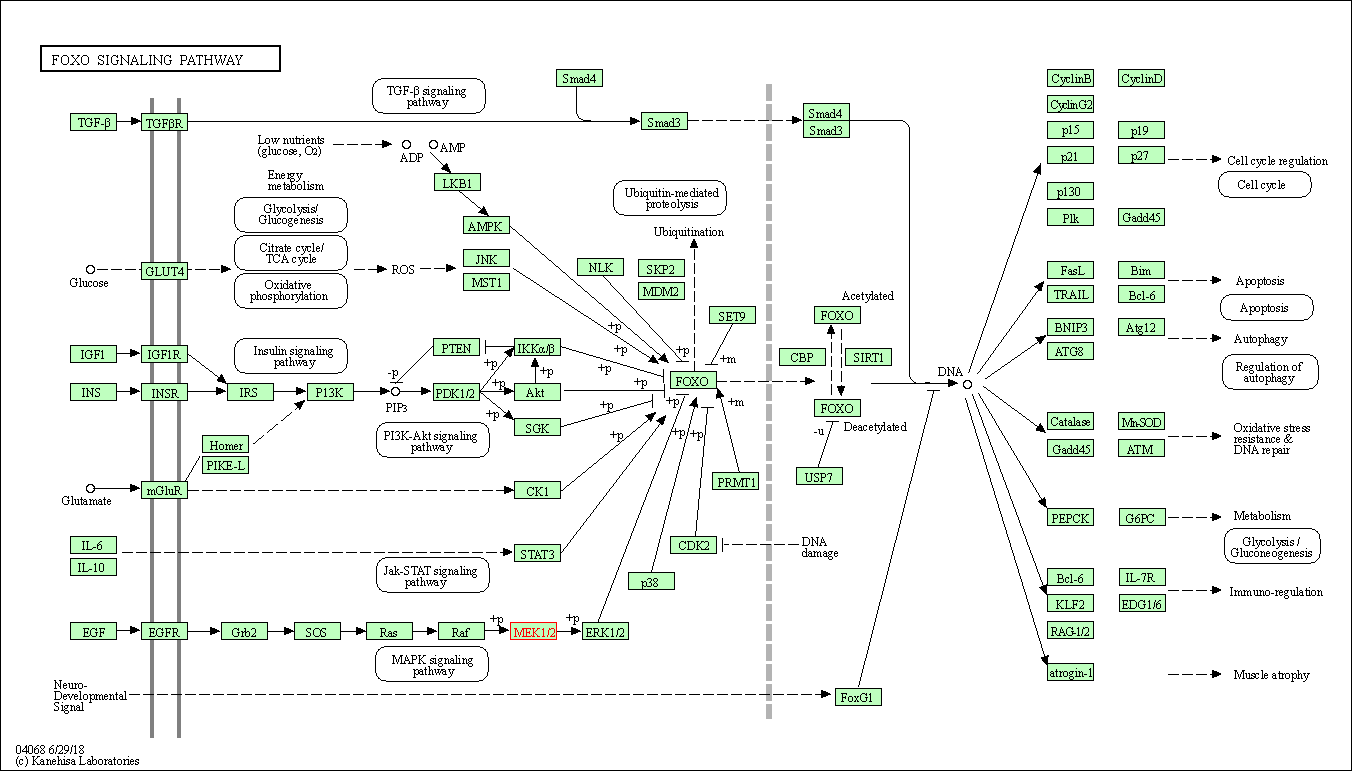
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |
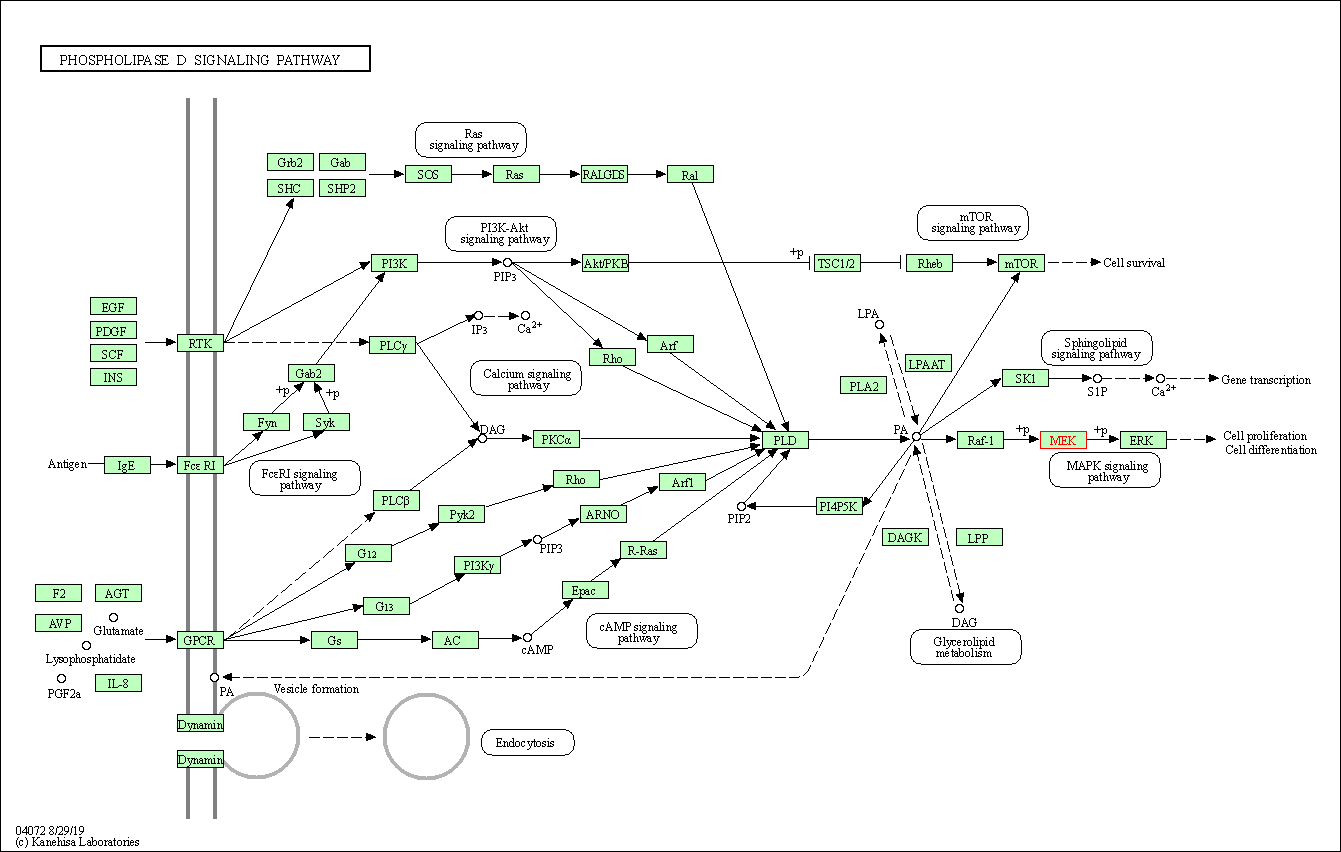
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| VEGF signaling pathway | hsa04370 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apelin signaling pathway | hsa04371 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Gap junction | hsa04540 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Neutrophil extracellular trap formation | hsa04613 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| B cell receptor signaling pathway | hsa04662 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Long-term depression | hsa04730 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Prolactin signaling pathway | hsa04917 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH secretion | hsa04929 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 14 | Degree centrality | 1.50E-03 | Betweenness centrality | 2.55E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.34E-01 | Radiality | 1.41E+01 | Clustering coefficient | 6.81E-01 |
| Neighborhood connectivity | 5.56E+01 | Topological coefficient | 1.61E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Drug Resistance Mutation (DRM) | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT01933932) Assess Efficacy & Safety of Selumetinib in Combination With Docetaxel in Patients Receiving 2nd Line Treatment for v-Ki-ras2 Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) Positive NSCLC (SELECT-1) | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800028258) | |||||
| REF 4 | Clinical pipeline report, company report or official report of MedKoo Biosciences. | |||||
| REF 5 | Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004 Dec;11(12):1192-7. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

