Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T63505
(Former ID: TTDNS00592)
|
|||||
| Target Name |
Tyrosine-protein kinase ABL1 (ABL)
|
|||||
| Synonyms |
p150; Proto-oncogene tyrosine-protein kinase ABL1; Proto-oncogene c-Abl; JTK7; C-ABL; Abl; Abelson tyrosine-protein kinase 1; Abelson murine leukemia viral oncogene homolog 1
Click to Show/Hide
|
|||||
| Gene Name |
ABL1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Breast cancer [ICD-11: 2C60-2C6Y] | |||||
| 2 | Ischemia [ICD-11: 8B10-8B11] | |||||
| 3 | Mature B-cell lymphoma [ICD-11: 2A85] | |||||
| 4 | Nutritional deficiency [ICD-11: 5B50-5B71] | |||||
| Function |
Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like WASF3 (involved in branch formation); ANXA1 (involved in membrane anchoring); DBN1, DBNL, CTTN, RAPH1 and ENAH (involved in signaling); or MAPT and PXN (microtubule-binding proteins). Phosphorylation of WASF3 is critical for the stimulation of lamellipodia formation and cell migration. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as BCAR1, CRK, CRKL, DOK1, EFS or NEDD9. Phosphorylates multiple receptor tyrosine kinases and more particularly promotes endocytosis of EGFR, facilitates the formation of neuromuscular synapses through MUSK, inhibits PDGFRB-mediated chemotaxis and modulates the endocytosis of activated B-cell receptor complexes. Other substrates which are involved in endocytosis regulation are the caveolin (CAV1) and RIN1. Moreover, ABL1 regulates the CBL family of ubiquitin ligases that drive receptor down-regulation and actin remodeling. Phosphorylation of CBL leads to increased EGFR stability. Involved in late-stage autophagy by regulating positively the trafficking and function of lysosomal components. ABL1 targets to mitochondria in response to oxidative stress and thereby mediates mitochondrial dysfunction and cell death. In response to oxidative stress, phosphorylates serine/threonine kinase PRKD2 at 'Tyr-717'. ABL1 is also translocated in the nucleus where it has DNA-binding activity and is involved in DNA-damage response and apoptosis. Many substrates are known mediators of DNA repair: DDB1, DDB2, ERCC3, ERCC6, RAD9A, RAD51, RAD52 or WRN. Activates the proapoptotic pathway when the DNA damage is too severe to be repaired. Phosphorylates TP73, a primary regulator for this type of damage-induced apoptosis. Phosphorylates the caspase CASP9 on 'Tyr-153' and regulates its processing in the apoptotic response to DNA damage. Phosphorylates PSMA7 that leads to an inhibition of proteasomal activity and cell cycle transition blocks. ABL1 acts also as a regulator of multiple pathological signaling cascades during infection. Several known tyrosine-phosphorylated microbial proteins have been identified as ABL1 substrates. This is the case of A36R of Vaccinia virus, Tir (translocated intimin receptor) of pathogenic E. coli and possibly Citrobacter, CagA (cytotoxin-associated gene A) of H. pylori, or AnkA (ankyrin repeat-containing protein A) of A. phagocytophilum. Pathogens can highjack ABL1 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1. Regulates T-cell differentiation in a TBX21-dependent manner. Phosphorylates TBX21 on tyrosine residues leading to an enhancement of its transcriptional activator activity. Non-receptor tyrosine-protein kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response and apoptosis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.10.2
|
|||||
| Sequence |
MLEICLKLVGCKSKKGLSSSSSCYLEEALQRPVASDFEPQGLSEAARWNSKENLLAGPSE
NDPNLFVALYDFVASGDNTLSITKGEKLRVLGYNHNGEWCEAQTKNGQGWVPSNYITPVN SLEKHSWYHGPVSRNAAEYLLSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTAS DGKLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSPNYDKWEMERT DITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQ LLGVCTREPPFYIITEFMTYGNLLDYLRECNRQEVNAVVLLYMATQISSAMEYLEKKNFI HRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLAYNKFSIKS DVWAFGVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNP SDRPSFAEIHQAFETMFQESSISDEVEKELGKQGVRGAVSTLLQAPELPTKTRTSRRAAE HRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERLLPKDKKTNLF SALIKKKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRDISNGALAFTPLDTADPAKSP KPSNGAGVPNGALRESGGSGFRSPHLWKKSSTLTSSRLATGEEEGGGSSSKRFLRSCSAS CVPHGAKDTEWRSVTLPRDLQSTGRQFDSSTFGGHKSEKPALPRKRAGENRSDQVTRGTV TPPPRLVKKNEEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGS ALGTPAAAEPVTPTSKAGSGAPGGTSKGPAEESRVRRHKHSSESPGRDKGKLSRLKPAPP PPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDAVNSDAAKPSQPGEGLKKPVL PATPKPQSAKPSGTPISPAPVPSTLPSASSALAGDQPSSTAFIPLISTRVSLRKTRQPPE RIASGAITKGVVLDSTEALCLAISRNSEQMASHSAVLEAGKNLYTFCVSYVDSIQQMRNK FAFREAINKLENNLRELQICPATAGSGPAATQDFSKLLSSVKEISDIVQR Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A00933 ; BADD_A02049 ; BADD_A07749 | |||||
| HIT2.0 ID | T75MO6 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 4 Approved Drugs | + | ||||
| 1 | Adenosine triphosphate | Drug Info | Approved | Malnutrition | [2] | |
| 2 | Bosutinib | Drug Info | Approved | Breast cancer | [3], [4] | |
| 3 | Ponatinib | Drug Info | Approved | Acute lymphoblastic leukaemia | [5], [6] | |
| 4 | SKI-758 | Drug Info | Approved | Ischemia | [6], [7] | |
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | Flumatinib | Drug Info | Phase 2 | Chronic myelogenous leukaemia | [10] | |
| 2 | DCC-2036 | Drug Info | Phase 1/2 | Chronic myeloid leukaemia | [11] | |
| 3 | ISIS-CRP | Drug Info | Phase 1 | Inflammation | [13] | |
| 4 | KW-2449 | Drug Info | Phase 1 | Acute myeloid leukaemia | [14], [15] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | MC-2001 | Drug Info | Preclinical | leukaemia | [16] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 34 Inhibitor drugs | + | ||||
| 1 | Adenosine triphosphate | Drug Info | [1], [17] | |||
| 2 | Bosutinib | Drug Info | [18] | |||
| 3 | SKI-758 | Drug Info | [19] | |||
| 4 | Flumatinib | Drug Info | [20] | |||
| 5 | ISIS-CRP | Drug Info | [22] | |||
| 6 | 6,6-fused nitrogenous heterocyclic compound 1 | Drug Info | [23] | |||
| 7 | 6,6-fused nitrogenous heterocyclic compound 2 | Drug Info | [23] | |||
| 8 | Azaindole derivative 2 | Drug Info | [23] | |||
| 9 | Indol-5-ol derivative 1 | Drug Info | [23] | |||
| 10 | PMID25656651-Compound-28b | Drug Info | [23] | |||
| 11 | PMID25656651-Compound-33a | Drug Info | [23] | |||
| 12 | PMID25656651-Compound-33b | Drug Info | [23] | |||
| 13 | PMID25656651-Compound-34a | Drug Info | [23] | |||
| 14 | PMID25656651-Compound-34b | Drug Info | [23] | |||
| 15 | PMID25656651-Compound-34c | Drug Info | [23] | |||
| 16 | PMID27774824-Compound-Figure9Example2down | Drug Info | [24] | |||
| 17 | MC-2001 | Drug Info | [25] | |||
| 18 | 4-[(3,5-diamino-1H-pyrazol-4-yl)diazenyl]phenol | Drug Info | [26] | |||
| 19 | AP-24226 | Drug Info | [27] | |||
| 20 | BAS-00387275 | Drug Info | [28] | |||
| 21 | BAS-00387328 | Drug Info | [28] | |||
| 22 | BAS-00387347 | Drug Info | [28] | |||
| 23 | BAS-00672722 | Drug Info | [28] | |||
| 24 | BAS-01373578 | Drug Info | [28] | |||
| 25 | BAS-0338872 | Drug Info | [29] | |||
| 26 | BAS-0338876 | Drug Info | [29] | |||
| 27 | BAS-09534324 | Drug Info | [28] | |||
| 28 | Bis-(5-hydroxy-1H-indol-2-yl)-methanone | Drug Info | [30] | |||
| 29 | MYRISTIC ACID | Drug Info | [31] | |||
| 30 | PD-0166326 | Drug Info | [32] | |||
| 31 | PD-0173956 | Drug Info | [32] | |||
| 32 | TG-100435 | Drug Info | [33] | |||
| 33 | TRISMETHOXYRESVERATROL | Drug Info | [34] | |||
| 34 | [1,1':2',1'']-terphenyl-4,3'',5''-triol | Drug Info | [34] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | Ponatinib | Drug Info | [7] | |||
| 2 | DCC-2036 | Drug Info | [21] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Nilotinib | Ligand Info | |||||
| Structure Description | ABL1 kinase (T334I_D382N) in complex with asciminib and nilotinib | PDB:5MO4 | ||||
| Method | X-ray diffraction | Resolution | 2.17 Å | Mutation | No | [35] |
| PDB Sequence |
NLFVALYDFV
92 ASGDNTLSIT102 KGEKLRVLGY112 NHNGEWCEAQ122 TKNGQGWVPS132 NYITPVNSLE 142 KHSWYHGPVS152 RNAAEYLLSS162 GINGSFLVRE172 SESSPGQRSI182 SLRYEGRVYH 192 YRINTASDGK202 LYVSSESRFN212 TLAELVHHHS222 TVADGLITTL232 HYPAPKRNKP 242 TVYGVSPNYD252 KWEMERTDIT262 MKHKLGGGQY272 GEVYEGVWKK282 YSLTVAVKTL 292 KEDEVEEFLK304 EAAVMKEIKH314 PNLVQLLGVC324 TREPPFYIII334 EFMTYGNLLD 344 YLRECNRQEV354 NAVVLLYMAT364 QISSAMEYLE374 KKNFIHRNLA384 ARNCLVGENH 394 LVKVADFFPI422 KWTAPESLAY432 NKFSIKSDVW442 AFGVLLWEIA452 TYGMSPYPGI 462 DLSQVYELLE472 KDYRMERPEG482 CPEKVYELMR492 ACWQWNPSDR502 PSFAEIHQAF 512 ETMFQESSIS522 DEVEKELGK
|
|||||
|
|
LEU267
3.707
TYR272
3.546
VAL275
3.810
ALA288
3.293
VAL289
3.982
LYS290
3.387
GLU301
4.988
LYS304
3.834
GLU305
2.948
VAL308
3.630
MET309
3.467
ILE312
3.637
LEU317
3.548
VAL318
3.571
ILE332
3.487
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Imatinib | Ligand Info | |||||
| Structure Description | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | PDB:3PYY | ||||
| Method | X-ray diffraction | Resolution | 1.85 Å | Mutation | No | [36] |
| PDB Sequence |
YDKWEMERTD
260 ITMKHKLGGG270 QYGEVYEGVW280 KKYSLTVAVK290 TLKEVEEFLK304 EAAVMKEIKH 314 PNLVQLLGVC324 TREPPFYIIT334 EFMTYGNLLD344 YLRECNRQEV354 NAVVLLYMAT 364 QISSAMEYLE374 KKNFIHRDLA384 ARNCLVGENH394 LVKVADFGLS404 RLMTGDTYTA 414 HAGAKFPIKW424 TAPESLAYNK434 FSIKSDVWAF444 GVLLWEIATY454 GMSPYPGIDL 464 SQVYELLEKD474 YRMERPEGCP484 EKVYELMRAC494 WQWNPSDRPS504 FAEIHQAFET 514 MFQES
|
|||||
|
|
LEU267
3.758
TYR272
3.815
VAL275
3.855
ALA288
3.508
VAL289
4.125
LYS290
3.642
GLU305
2.942
VAL308
3.876
MET309
3.439
ILE312
3.655
VAL318
3.110
ILE332
3.752
THR334
2.978
GLU335
4.418
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| ErbB signaling pathway | hsa04012 | Affiliated Target |
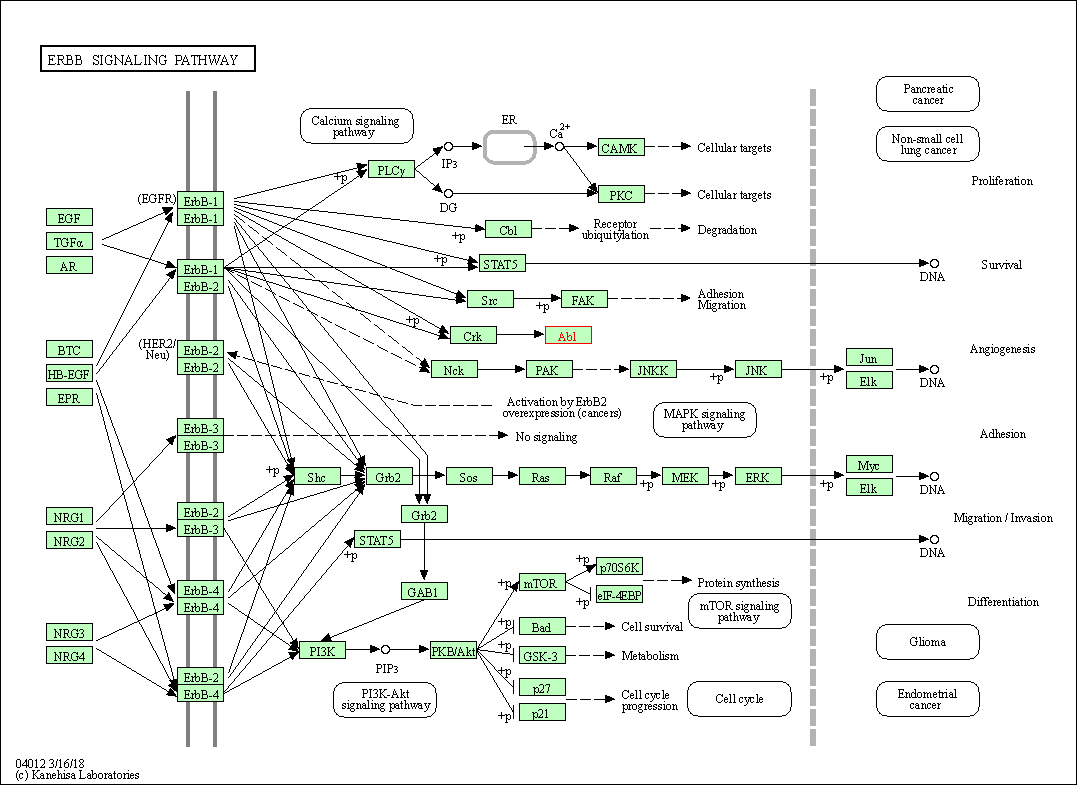
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |
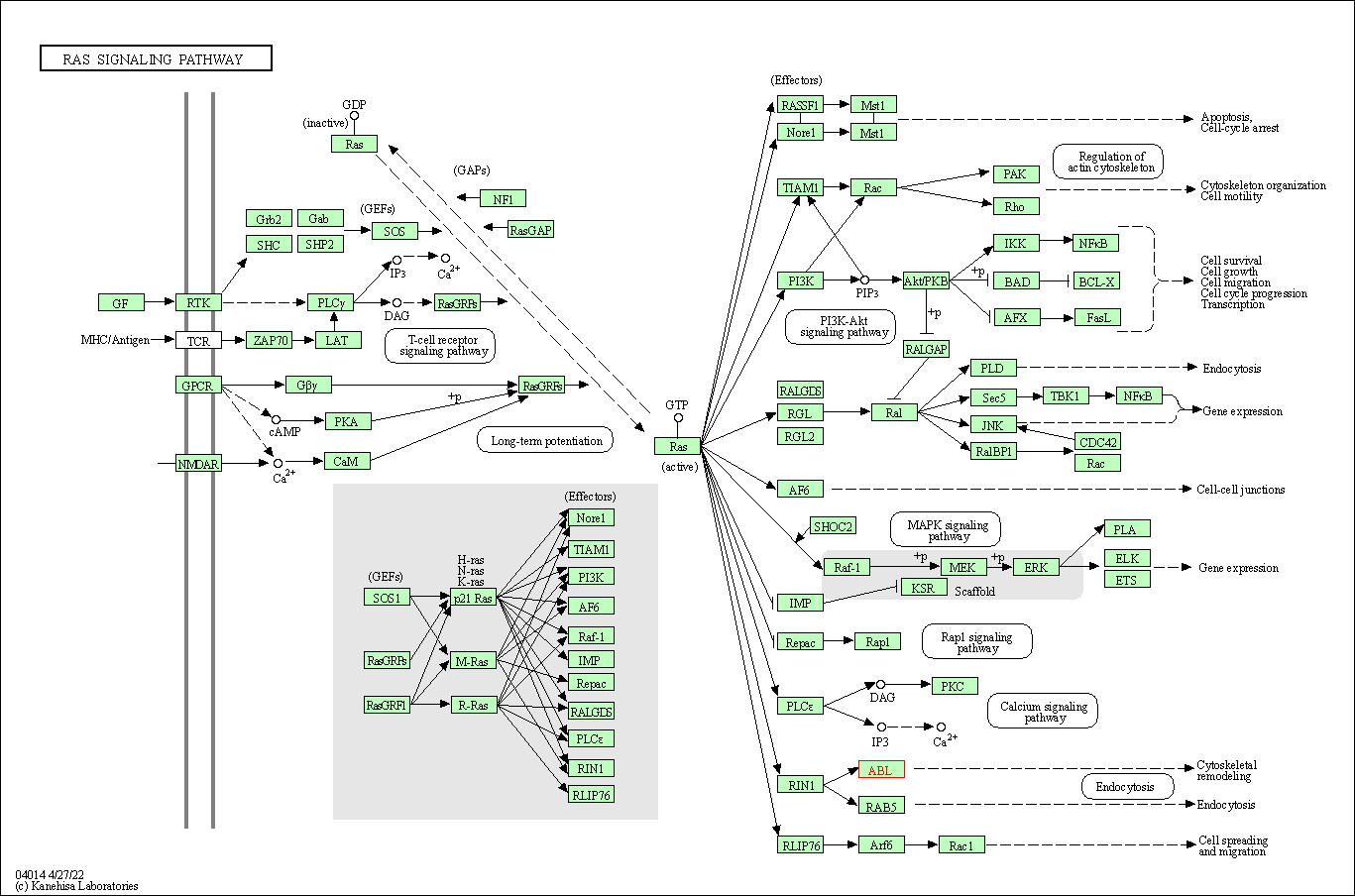
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cell cycle | hsa04110 | Affiliated Target |
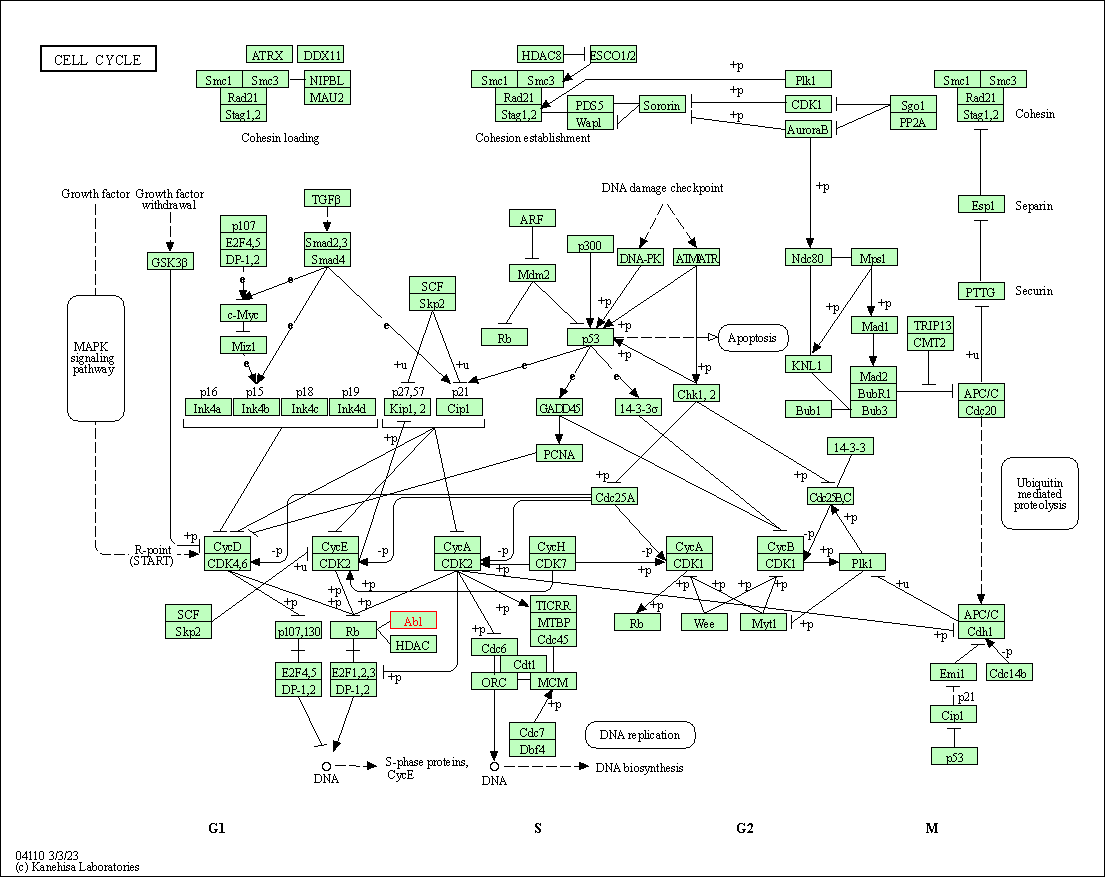
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Axon guidance | hsa04360 | Affiliated Target |
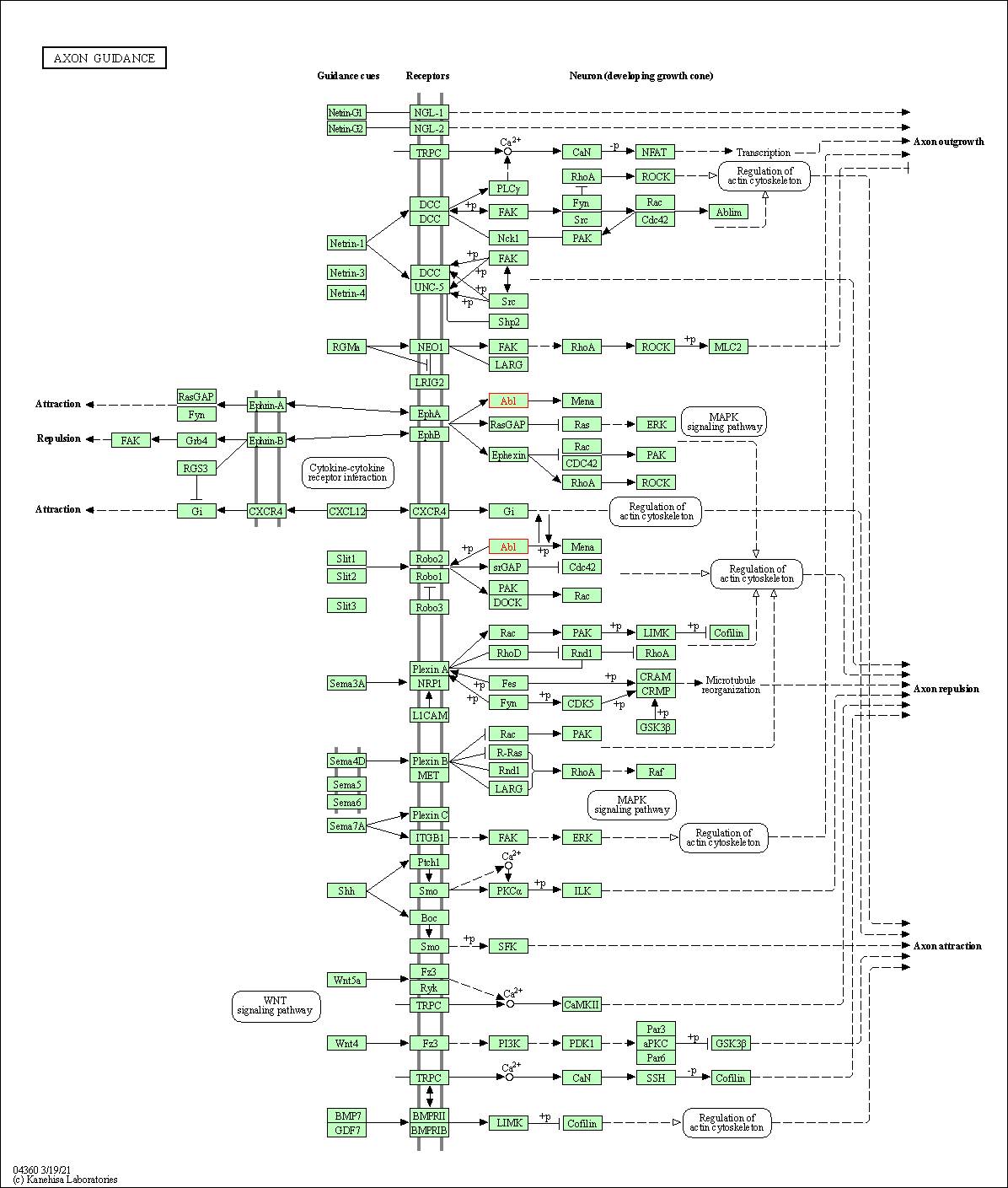
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Degree | 38 | Degree centrality | 4.08E-03 | Betweenness centrality | 3.00E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.64E-01 | Radiality | 1.45E+01 | Clustering coefficient | 1.22E-01 |
| Neighborhood connectivity | 5.31E+01 | Topological coefficient | 5.34E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Drug Resistance Mutation (DRM) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Targeted chronic myeloid leukemia therapy: Seeking a cure. Am J Health Syst Pharm. 2007 Dec 15;64(24 Suppl 15):S9-15. | |||||
| REF 2 | Emerging drugs for chemotherapy-induced mucositis. Expert Opin Emerg Drugs. 2008 Sep;13(3):511-22. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5710). | |||||
| REF 4 | ClinicalTrials.gov (NCT02311998) Phase I/II Study of Bosutinib in Combination With Inotuzumab Ozogamicin in CD22-positive Philadelphia-Chromosome (PC) Positive Acute Lymphoblastic Leukemia (ALL) and Chronic Myeloid Leukemia (CML). U.S. National Institutes of Health. | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5890). | |||||
| REF 6 | Nat Rev Drug Discov. 2013 Feb;12(2):87-90. | |||||
| REF 7 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 8 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7731). | |||||
| REF 9 | Current and future treatments of bone metastases. Expert Opin Emerg Drugs. 2008 Dec;13(4):609-27. | |||||
| REF 10 | ClinicalTrials.gov (NCT02511340) A Phase II Study to Evaluate the Efficacy and the Safety of Flumatinib in CML-AP or CML-BP Patients. | |||||
| REF 11 | ClinicalTrials.gov (NCT00827138) Study Safety and Preliminary Efficacy of DCC-2036 in Patients With Leukemias (Ph+ CML With T315I Mutation). U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT04350177) A Phase I, Randomized Single Ascending Dose (SAD) and Multiple Ascending Dose (MAD) Study to Determine the Safety, Tolerability and Pharmacokinetics (PK) of IkT-148009 in Older Adult and Elderly Healthy Volunteers With Extension Into Parkinson's Patients. U.S.National Institutes of Health. | |||||
| REF 13 | Clinical pipeline report, company report or official report of ISIS Pharmaceuticals (2011). | |||||
| REF 14 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5691). | |||||
| REF 15 | KW-2449, a novel multikinase inhibitor, suppresses the growth of leukemia cells with FLT3 mutations or T315I-mutated BCR/ABL translocation. Blood. 2009 Aug 20;114(8):1607-17. | |||||
| REF 16 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800021088) | |||||
| REF 17 | High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002 May 1;99(9):3472-5. | |||||
| REF 18 | A comparison of physicochemical property profiles of marketed oral drugs and orally bioavailable anti-cancer protein kinase inhibitors in clinical development. Curr Top Med Chem. 2007;7(14):1408-22. | |||||
| REF 19 | Synthesis and Src kinase inhibitory activity of a series of 4-[(2,4-dichloro-5-methoxyphenyl)amino]-7-furyl-3-quinolinecarbonitriles. J Med Chem. 2006 Dec 28;49(26):7868-76. | |||||
| REF 20 | Flumatinib, a selective inhibitor of BCR-ABL/PDGFR/KIT, effectively overcomes drug resistance of certain KIT mutants. Cancer Sci. 2014 Jan;105(1):117-25. | |||||
| REF 21 | Company report (Deciphera Pharmaceuticals: Tumor-Targeted Programs and Indications) | |||||
| REF 22 | Structure-based optimization of pyrazolo[3,4-d]pyrimidines as Abl inhibitors and antiproliferative agents toward human leukemia cell lines. J Med Chem. 2008 Mar 13;51(5):1252-9. | |||||
| REF 23 | Bcr-Abl tyrosine kinase inhibitors: a patent review.Expert Opin Ther Pat. 2015 Apr;25(4):397-412. | |||||
| REF 24 | Inhibitors of JAK-family kinases: an update on the patent literature 2013-2015, part 1.Expert Opin Ther Pat. 2017 Feb;27(2):127-143. | |||||
| REF 25 | Vascular endothelial growth factor and its receptors in multiple myeloma. Leukemia. 2003 Oct;17(10):1961-6. | |||||
| REF 26 | 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J Med Chem. 2006 Nov 2;49(22):6500-9. | |||||
| REF 27 | Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP2453... J Med Chem. 2010 Jun 24;53(12):4701-19. | |||||
| REF 28 | A combination of docking/dynamics simulations and pharmacophoric modeling to discover new dual c-Src/Abl kinase inhibitors. J Med Chem. 2006 Jun 1;49(11):3278-86. | |||||
| REF 29 | Discovery and SAR of 1,3,4-thiadiazole derivatives as potent Abl tyrosine kinase inhibitors and cytodifferentiating agents. Bioorg Med Chem Lett. 2008 Feb 1;18(3):1207-11. | |||||
| REF 30 | Novel bis(1H-indol-2-yl)methanones as potent inhibitors of FLT3 and platelet-derived growth factor receptor tyrosine kinase. J Med Chem. 2006 Jun 1;49(11):3101-15. | |||||
| REF 31 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 32 | Structure-activity relationships of 6-(2,6-dichlorophenyl)-8-methyl-2-(phenylamino)pyrido[2,3-d]pyrimidin-7-ones: toward selective Abl inhibitors. Bioorg Med Chem Lett. 2009 Dec 15;19(24):6872-6. | |||||
| REF 33 | Discovery of [7-(2,6-dichlorophenyl)-5-methylbenzo [1,2,4]triazin-3-yl]-[4-(2-pyrrolidin-1-ylethoxy)phenyl]amine--a potent, orally active Src kinas... Bioorg Med Chem Lett. 2007 Feb 1;17(3):602-8. | |||||
| REF 34 | Identification of a terphenyl derivative that blocks the cell cycle in the G0-G1 phase and induces differentiation in leukemia cells. J Med Chem. 2006 May 18;49(10):3012-8. | |||||
| REF 35 | The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature. 2017 Mar 30;543(7647):733-737. | |||||
| REF 36 | Discovery and characterization of a cell-permeable, small-molecule c-Abl kinase activator that binds to the myristoyl binding site. Chem Biol. 2011 Feb 25;18(2):177-86. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

