Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T01908
(Former ID: TTDC00028)
|
|||||
| Target Name |
Tissue kallikrein (KLK2)
|
|||||
| Synonyms |
hGK-1; Tissue kallikrein-2; Kallikrein-2; Glandular kallikrein-1
Click to Show/Hide
|
|||||
| Gene Name |
KLK2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Innate/adaptive immunodeficiency [ICD-11: 4A00] | |||||
| Function |
Glandular kallikreins cleave Met-Lys and Arg-Ser bonds in kininogen to release Lys-bradykinin.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.21.35
|
|||||
| Sequence |
MWDLVLSIALSVGCTGAVPLIQSRIVGGWECEKHSQPWQVAVYSHGWAHCGGVLVHPQWV
LTAAHCLKKNSQVWLGRHNLFEPEDTGQRVPVSHSFPHPLYNMSLLKHQSLRPDEDSSHD LMLLRLSEPAKITDVVKVLGLPTQEPALGTTCYASGWGSIEPEEFLRPRSLQCVSLHLLS NDMCARAYSEKVTEFMLCAGLWTGGKDTCGGDSGGPLVCNGVLQGITSWGPEPCALPEKP AVYTKVVHYRKWIKDTIAANP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 5 Clinical Trial Drugs | + | ||||
| 1 | Avoralstat | Drug Info | Phase 2/3 | Hereditary angioedema | [2] | |
| 2 | VDA-1102 | Drug Info | Phase 2 | Actinic keratosis | [3] | |
| 3 | JNJ-69086420 | Drug Info | Phase 1 | Prostate cancer | [4] | |
| 4 | JNJ-75229414 | Drug Info | Phase 1 | Prostate cancer | [5] | |
| 5 | JNJ-78278343 | Drug Info | Phase 1 | Prostate cancer | [6] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | Dermolastin | Drug Info | Discontinued in Phase 2 | Atopic dermatitis | [7] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Avoralstat | Drug Info | [1] | |||
| 2 | JNJ-69086420 | Drug Info | [4] | |||
| 3 | Dermolastin | Drug Info | [9], [10], [11], [12] | |||
| 4 | DM-107 | Drug Info | [13] | |||
| 5 | hK2p01 derivative KLK2 inhibitor | Drug Info | [14] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | VDA-1102 | Drug Info | [8] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Benzamidine | Ligand Info | |||||
| Structure Description | Human kallikrein-related peptidase 2 in complex with benzamidine | PDB:4NFE | ||||
| Method | X-ray diffraction | Resolution | 1.90 Å | Mutation | No | [15] |
| PDB Sequence |
IVGGWECEKH
25 SQPWQVAVYS35 HGWAHCGGVL46 VHPQWVLTAA56 HCLKKNSQVW67 LGRHNLFEPE 77 DTGQRVPVSH87 SFPHPLYNMS95B LDSSHDLMLL106 RLSEPAKITD116 VVKVLGLPTQ 128 EPALGTTCYA138 SGWGSIEPEE148 FLRPRSLQCV158 SLHLLSNDMC168 ARAYSEKVTE 178 FMLCAGLWTG186B GKDTCGGDSG196 GPLVCNGVLQ210 GITSWGPEPC220 ALPEKPAVYT 229 KVVHYRKWIK239 DTIAANP
|
|||||
|
|
TRP20
2.934
GLU21
4.169
CYS22
3.583
GLU23
2.956
SER26
3.723
GLN27
3.790
TYR137
2.542
CYS157
4.376
ASP189
2.812
THR190
2.815
CYS191
3.896
GLY192
3.677
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Renin-angiotensin system | hsa04614 | Affiliated Target |
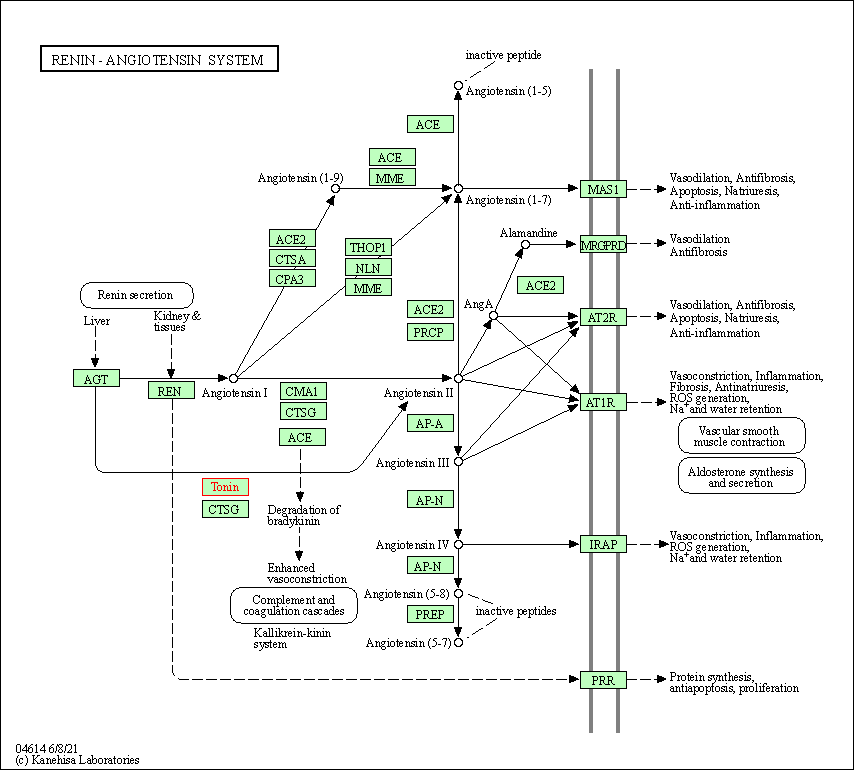
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Endocrine and other factor-regulated calcium reabsorption | hsa04961 | Affiliated Target |
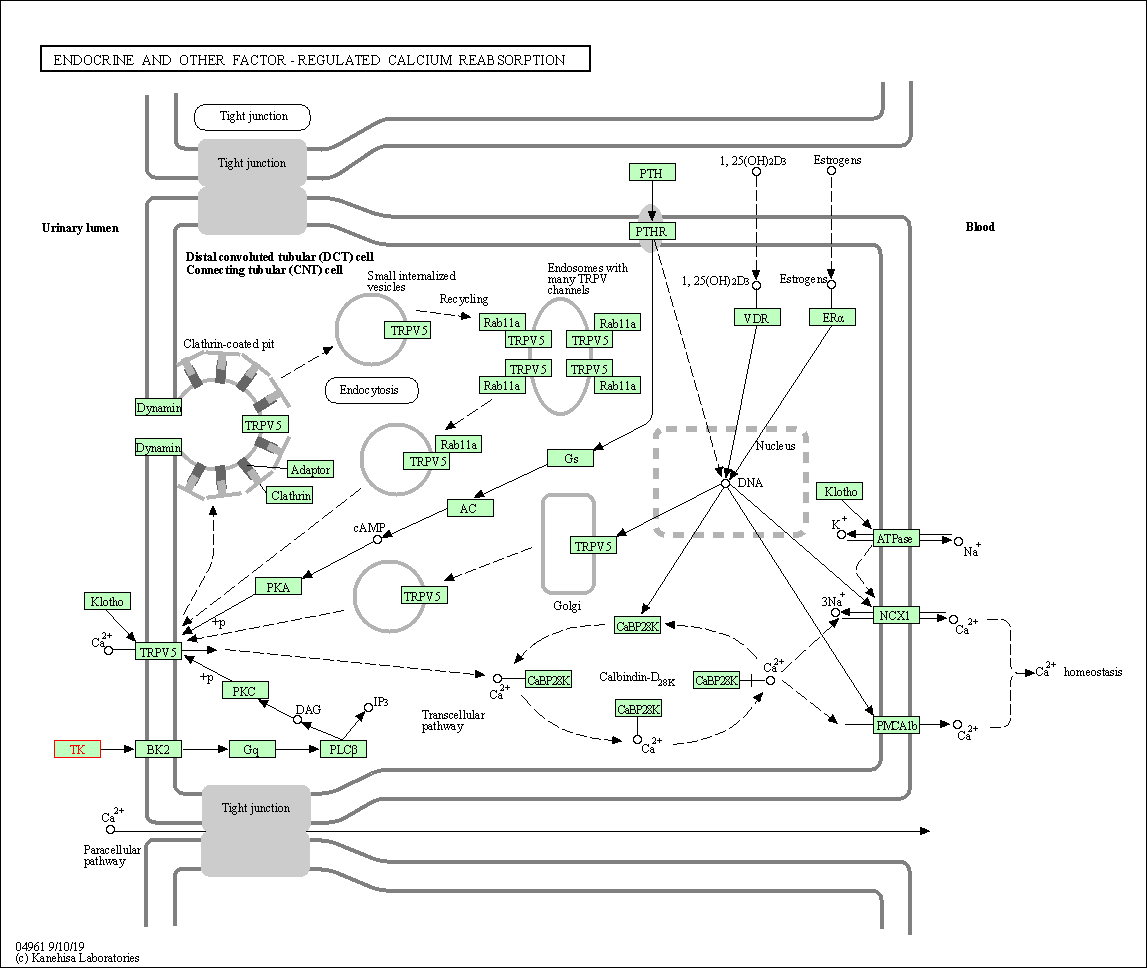
|
| Class: Organismal Systems => Excretory system | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 2.13E-01 | Radiality | 1.37E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 6.10E+01 | Topological coefficient | 1.00E+00 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Endocrine and other factor-regulated calcium reabsorption | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | IL2 Signaling Pathway | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | Coregulation of Androgen receptor activity | |||||
| 2 | Regulation of Androgen receptor activity | |||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | Activation of Matrix Metalloproteinases | |||||
| 2 | Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | |||||
| 3 | Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Prostate Cancer | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT02303626) 12-Week Safety and Efficacy Study of BCX4161 as an Oral Prophylaxis Against HAE Attacks. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT03538951) Study to Evaluate the Efficacy, Safety, and Tolerability of Topical VDA-1102 Ointment in Subjects With Actinic Keratosis (Phase2b). U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT04644770) A Study of JNJ-69086420, an Actinium-225-Labeled Antibody Targeting Human Kallikrein-2 (hK2) for Advanced Prostate Cancer. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT05022849) A Phase 1, Dose Escalation Study of JNJ-75229414, a Chimeric Antigen Receptor T Cell (CAR-T) Therapy Directed Against KLK2 for Metastatic Castration-Resistant Prostate Cancer. U.S.National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT04898634) A Phase 1 Study of JNJ-78278343, a T-Cell-Redirecting Agent Targeting Human Kallikrein 2 (KLK2), for Advanced Prostate Cancer. U.S.National Institutes of Health. | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800018071) | |||||
| REF 8 | Clinical pipeline report, company report or official report of Vidac Pharma. | |||||
| REF 9 | Arriva-ProMetic recombinant alpha 1-antitrypsin (rAAT) moves into the clinic for dermatology applications. ProMetic Life Sciences. 2009. | |||||
| REF 10 | rAAt (inhaled) Arriva/Hyland Immuno. Curr Opin Mol Ther. 2006 Feb;8(1):76-82. | |||||
| REF 11 | Optimization of the bioprocessing conditions for scale-up of transient production of a heterologous protein in plants using a chemically inducible viral amplicon expression system. Biotechnol Prog. 2009 May-Jun;25(3):722-34. | |||||
| REF 12 | Bioreactor strategies for improving production yield and functionality of a recombinant human protein in transgenic tobacco cell cultures. Biotechnol Bioeng. 2009 Feb 1;102(2):508-20. | |||||
| REF 13 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2372). | |||||
| REF 14 | Novel peptide inhibitors of human kallikrein 2. J Biol Chem. 2006 May 5;281(18):12555-60. | |||||
| REF 15 | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity. J Biol Chem. 2014 Dec 5;289(49):34267-83. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

