Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T15051
(Former ID: TTDI00232)
|
|||||
| Target Name |
TNF receptor-associated factor 6 (TRAF6)
|
|||||
| Synonyms |
RNF85; RING-type E3 ubiquitin transferase TRAF6; RING finger protein 85; Interleukin-1 signal transducer; E3 ubiquitin-protein ligase TRAF6
Click to Show/Hide
|
|||||
| Gene Name |
TRAF6
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Mediates ubiquitination of free/unanchored polyubiquitin chain that leads to MAP3K7 activation. Leads to the activation of NF-kappa-B and JUN. May be essential for the formation of functional osteoclasts. Seems to also play a role in dendritic cells (DCs) maturation and/or activation. Represses c-Myb-mediated transactivation, in B-lymphocytes. Adapter protein that seems to play a role in signal transduction initiated via TNF receptor, IL-1 receptor and IL-17 receptor. Regulates osteoclast differentiation by mediating the activation of adapter protein complex 1 (AP-1) and NF-kappa-B, in response to RANK-L stimulation. Together with MAP3K8, mediates CD40 signals that activate ERK in B-cells and macrophages, and thus may play a role in the regulation of immunoglobulin production. E3 ubiquitin ligase that, together with UBE2N and UBE2V1, mediates the synthesis of 'Lys-63'-linked-polyubiquitin chains conjugated to proteins, such as IKBKG, IRAK1, AKT1 and AKT2.
Click to Show/Hide
|
|||||
| BioChemical Class |
Acyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.3.2.27
|
|||||
| Sequence |
MSLLNCENSCGSSQSESDCCVAMASSCSAVTKDDSVGGTASTGNLSSSFMEEIQGYDVEF
DPPLESKYECPICLMALREAVQTPCGHRFCKACIIKSIRDAGHKCPVDNEILLENQLFPD NFAKREILSLMVKCPNEGCLHKMELRHLEDHQAHCEFALMDCPQCQRPFQKFHINIHILK DCPRRQVSCDNCAASMAFEDKEIHDQNCPLANVICEYCNTILIREQMPNHYDLDCPTAPI PCTFSTFGCHEKMQRNHLARHLQENTQSHMRMLAQAVHSLSVIPDSGYISEVRNFQETIH QLEGRLVRQDHQIRELTAKMETQSMYVSELKRTIRTLEDKVAEIEAQQCNGIYIWKIGNF GMHLKCQEEEKPVVIHSPGFYTGKPGYKLCMRLHLQLPTAQRCANYISLFVHTMQGEYDS HLPWPFQGTIRLTILDQSEAPVRQNHEEIMDAKPELLAFQRPTIPRNPKGFGYVTFMHLE ALRQRTFIKDDTLLVRCEVSTRFDMGSLRREGFQPRSTDAGV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T76NQY | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
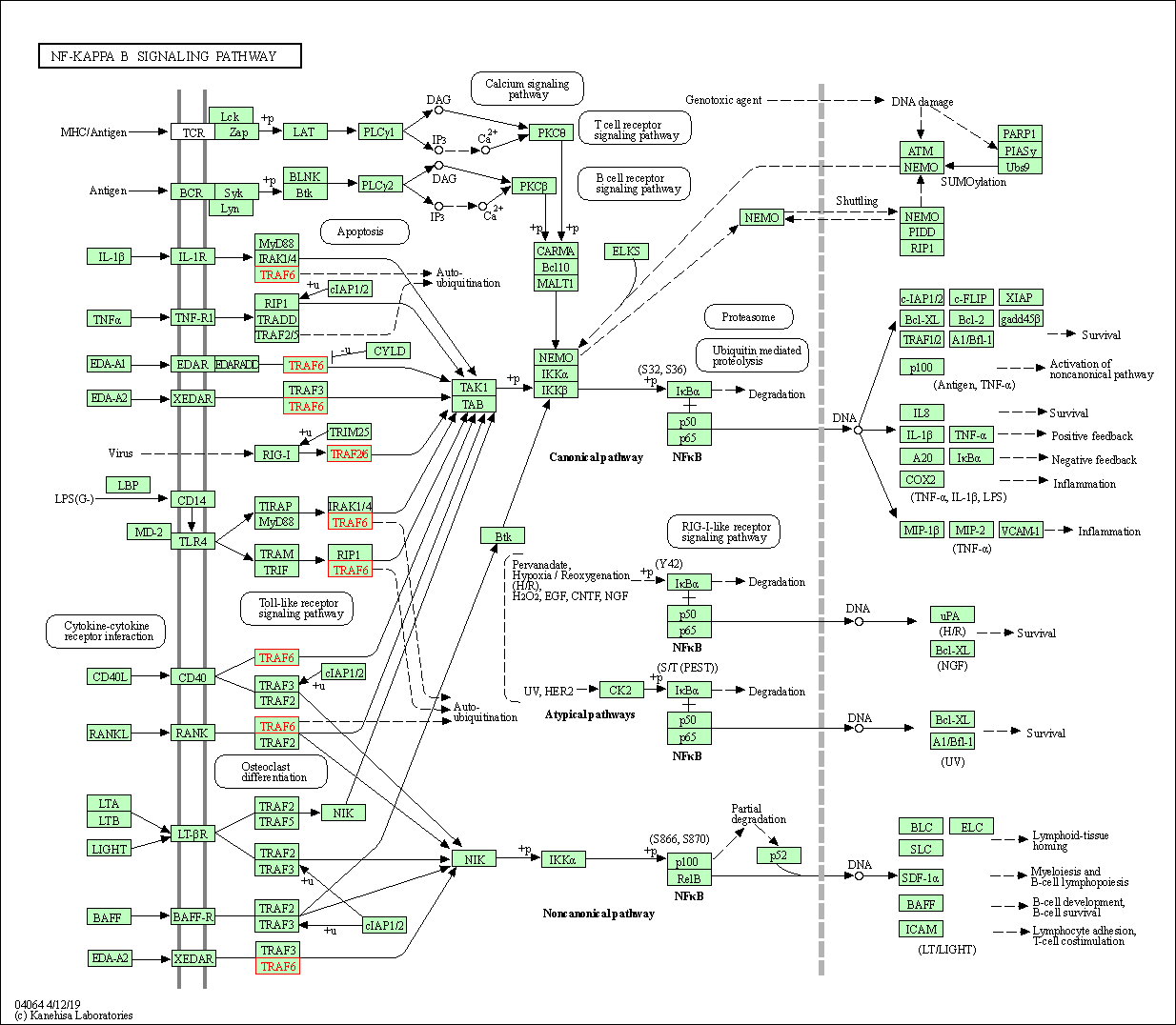
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
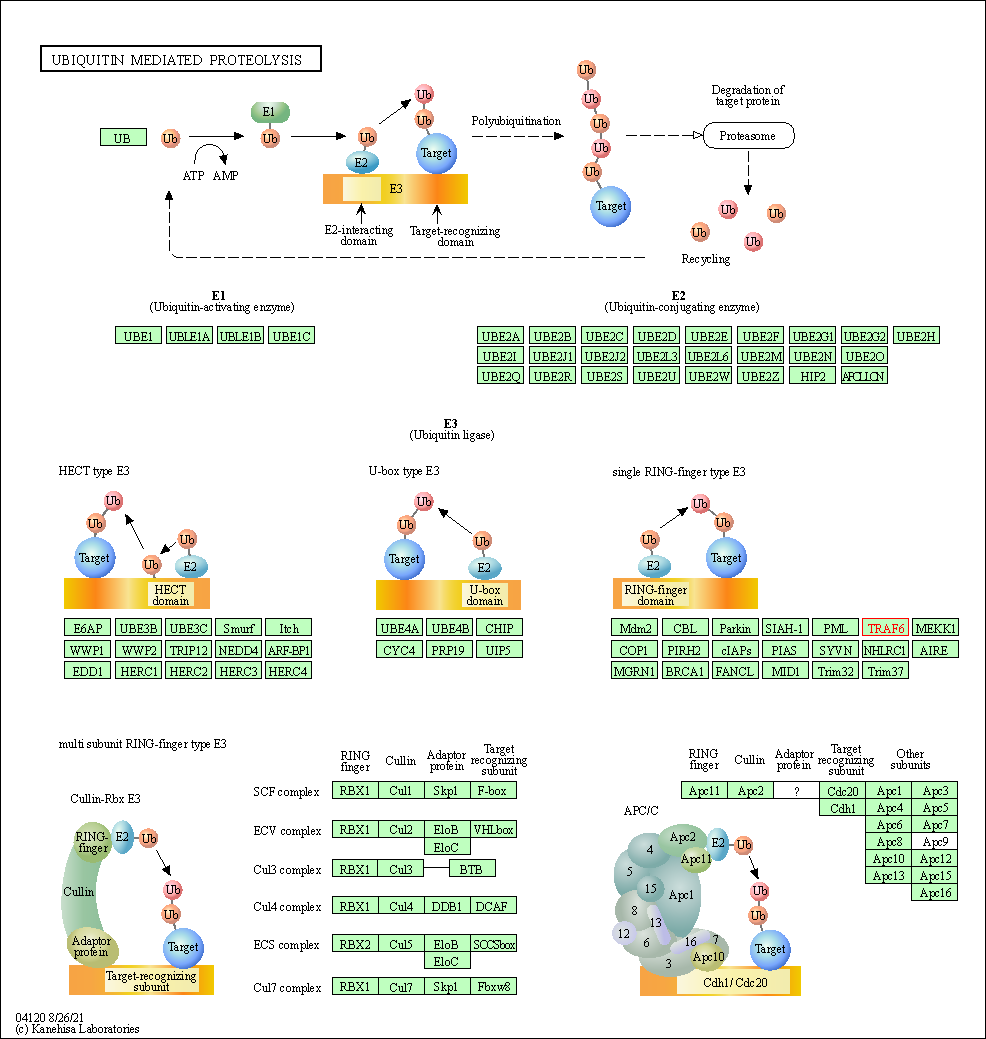
| Ubiquitin mediated proteolysis | hsa04120 | Affiliated Target |
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
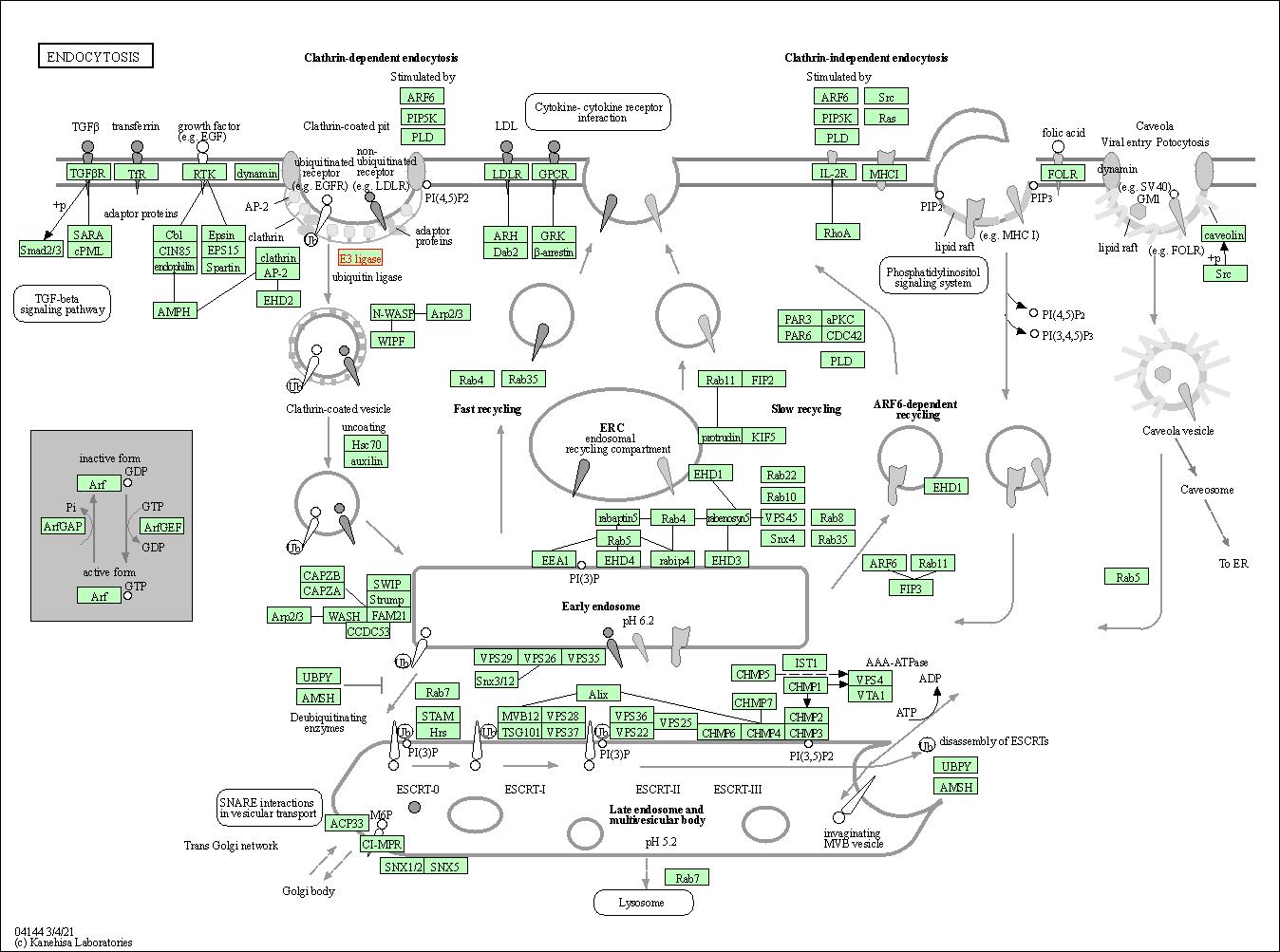
| Endocytosis | hsa04144 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
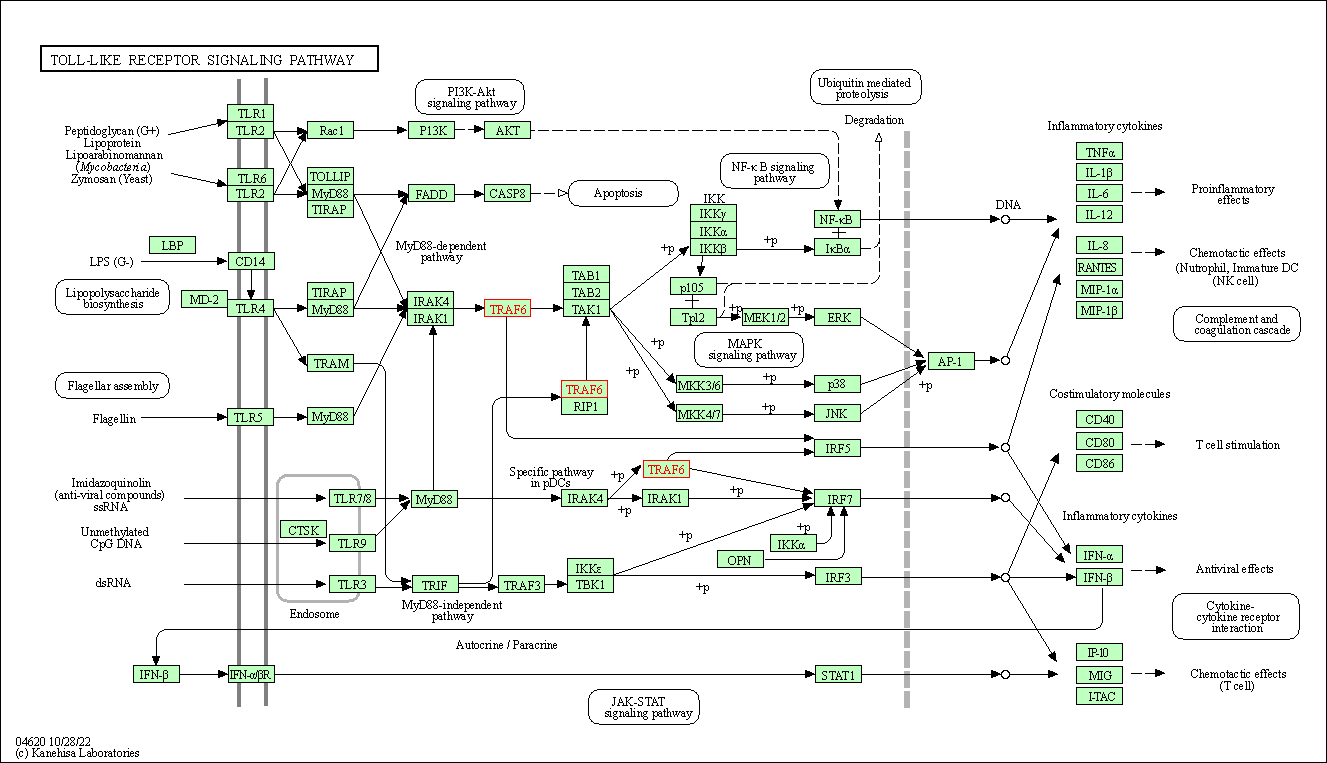
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| RIG-I-like receptor signaling pathway | hsa04622 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 97 | Degree centrality | 1.04E-02 | Betweenness centrality | 1.42E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.75E-01 | Radiality | 1.47E+01 | Clustering coefficient | 1.02E-01 |
| Neighborhood connectivity | 3.38E+01 | Topological coefficient | 2.40E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Tumor necrosis factor receptor-associated factor 6 as a nuclear factor kappa B-modulating therapeutic target in cardiovascular diseases: at the hea... Transl Res. 2018 May;195:48-61. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

