Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T17345
(Former ID: TTDI00931)
|
|||||
| Target Name |
Dihydrofolate reductase (DHFR)
|
|||||
| Synonyms |
DYR; DHFRP1
Click to Show/Hide
|
|||||
| Gene Name |
DHFR
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Indeterminate colitis [ICD-11: DD72] | |||||
| 2 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| 3 | Tuberculosis [ICD-11: 1B10-1B12] | |||||
| 4 | Urinary tract infection [ICD-11: GC08] | |||||
| Function |
Contributes to the de novo mitochondrial thymidylate biosynthesis pathway. Catalyzes an essential reaction for de novo glycine and purine synthesis, and for DNA precursor synthesis. Binds its own mRNA and that of DHFR2. Key enzyme in folate metabolism.
Click to Show/Hide
|
|||||
| BioChemical Class |
CH-NH donor oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.5.1.3
|
|||||
| Sequence |
MVGSLNCIVAVSQNMGIGKNGDLPWPPLRNEFRYFQRMTTTSSVEGKQNLVIMGKKTWFS
IPEKNRPLKGRINLVLSRELKEPPQGAHFLSRSLDDALKLTEQPELANKVDMVWIVGGSS VYKEAMNHPGHLKLFVTRIMQDFESDTFFPEIDLEKYKLLPEYPGVLSDVQEEKGIKYKF EVYEKND Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T02P7Q | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 4 Approved Drugs | + | ||||
| 1 | Aminosalicylic Acid | Drug Info | Approved | Pulmonary and extrapulmonary tuberculosis | [2] | |
| 2 | Leucovorin Calcium | Drug Info | Approved | Solid tumour/cancer | [2] | |
| 3 | Methotrexate Sodium | Drug Info | Approved | Solid tumour/cancer | [2] | |
| 4 | Trimethoprim | Drug Info | Approved | Urinary tract infection | [3] | |
| Clinical Trial Drug(s) | [+] 5 Clinical Trial Drugs | + | ||||
| 1 | Iclaprim | Drug Info | Phase 3 | Bacterial infection | [4] | |
| 2 | CH-4051 | Drug Info | Phase 2 | Rheumatoid arthritis | [5] | |
| 3 | PIRITREXIM | Drug Info | Phase 2 | Bladder cancer | [6], [7] | |
| 4 | L-MDAM | Drug Info | Phase 1 | Solid tumour/cancer | [9] | |
| 5 | MDAM (y-methylene-10-deazaaminopterin) | Drug Info | Phase 1 | Solid tumour/cancer | [10] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | TNP-351 | Drug Info | Discontinued in Phase 2 | Solid tumour/cancer | [11] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | 1954U89 | Drug Info | Preclinical | Solid tumour/cancer | [12] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 6 Modulator drugs | + | ||||
| 1 | Aminosalicylic Acid | Drug Info | [1] | |||
| 2 | Leucovorin Calcium | Drug Info | [13] | |||
| 3 | Methotrexate Sodium | Drug Info | [1] | |||
| 4 | Trimethoprim | Drug Info | [1] | |||
| 5 | PIRITREXIM | Drug Info | [16] | |||
| 6 | 1954U89 | Drug Info | [12] | |||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Iclaprim | Drug Info | [14] | |||
| 2 | CH-4051 | Drug Info | [15] | |||
| 3 | L-MDAM | Drug Info | [17] | |||
| 4 | MDAM (y-methylene-10-deazaaminopterin) | Drug Info | [18] | |||
| 5 | TNP-351 | Drug Info | [19] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Folic acid | Ligand Info | |||||
| Structure Description | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND A LIPOPHILIC ANTIFOLATE SELECTIVE FOR MYCOBACTERIUM AVIUM DHFR, 6-((2,5- DIETHOXYPHENYL)AMINOMETHYL)-2,4-DIAMINO-5-METHYLPYRIDO(2,3-D) PYRIMIDINE (SRI-8686) | PDB:2W3B | ||||
| Method | X-ray diffraction | Resolution | 1.27 Å | Mutation | No | [20] |
| PDB Sequence |
VGSLNCIVAV
10 SQNMGIGKNG20 DLPWPPLRNE30 FRYFQRMTTT40 SSVEGKQNLV50 IMGKKTWFSI 60 PEKNRPLKGR70 INLVLSRELK80 EPPQGAHFLS90 RSLDDALKLT100 EQPELANKVD 110 MVWIVGGSSV120 YKEAMNHPGH130 LKLFVTRIMQ140 DFESDTFFPE150 IDLEKYKLLP 160 EYPGVLSDVQ170 EEKGIKYKFE180 VYEKN
|
|||||
|
|
ILE7
3.363
VAL8
3.620
ALA9
3.609
LEU22
3.934
ARG28
4.531
GLU30
2.533
PHE31
3.622
ARG32
3.615
TYR33
4.502
PHE34
3.351
GLN35
0.860
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Trimethoprim | Ligand Info | |||||
| Structure Description | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND TRIMETHOPRIM | PDB:2W3A | ||||
| Method | X-ray diffraction | Resolution | 1.50 Å | Mutation | No | [21] |
| PDB Sequence |
VGSLNCIVAV
10 SQNMGIGKNG20 DLPWPPLRNE30 FRYFQRMTTT40 SSVEGKQNLV50 IMGKKTWFSI 60 PEKNRPLKGR70 INLVLSRELK80 EPPQGAHFLS90 RSLDDALKLT100 EQPELANKVD 110 MVWIVGGSSV120 YKEAMNHPGH130 LKLFVTRIMQ140 DFESDTFFPE150 IDLEKYKLLP 160 EYPGVLSDVQ170 EEKGIKYKFE180 VYEKND
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| One carbon pool by folate | hsa00670 | Affiliated Target |
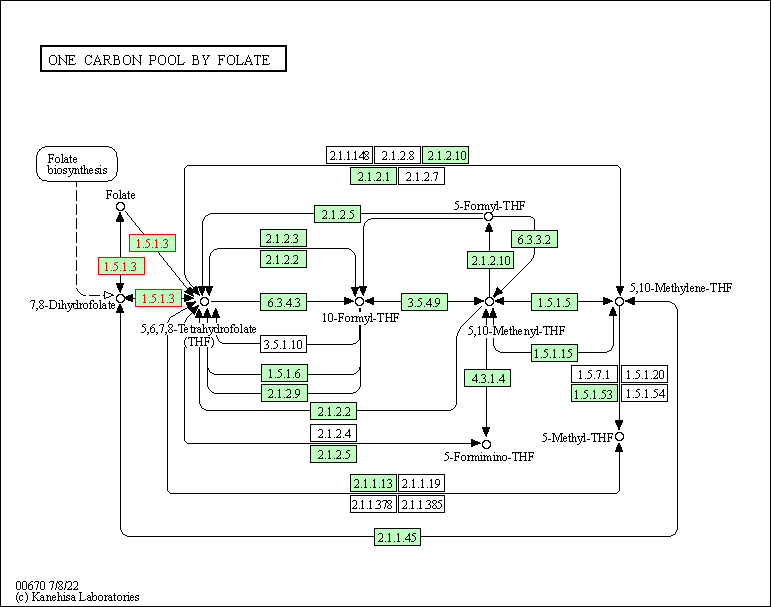
|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Folate biosynthesis | hsa00790 | Affiliated Target |
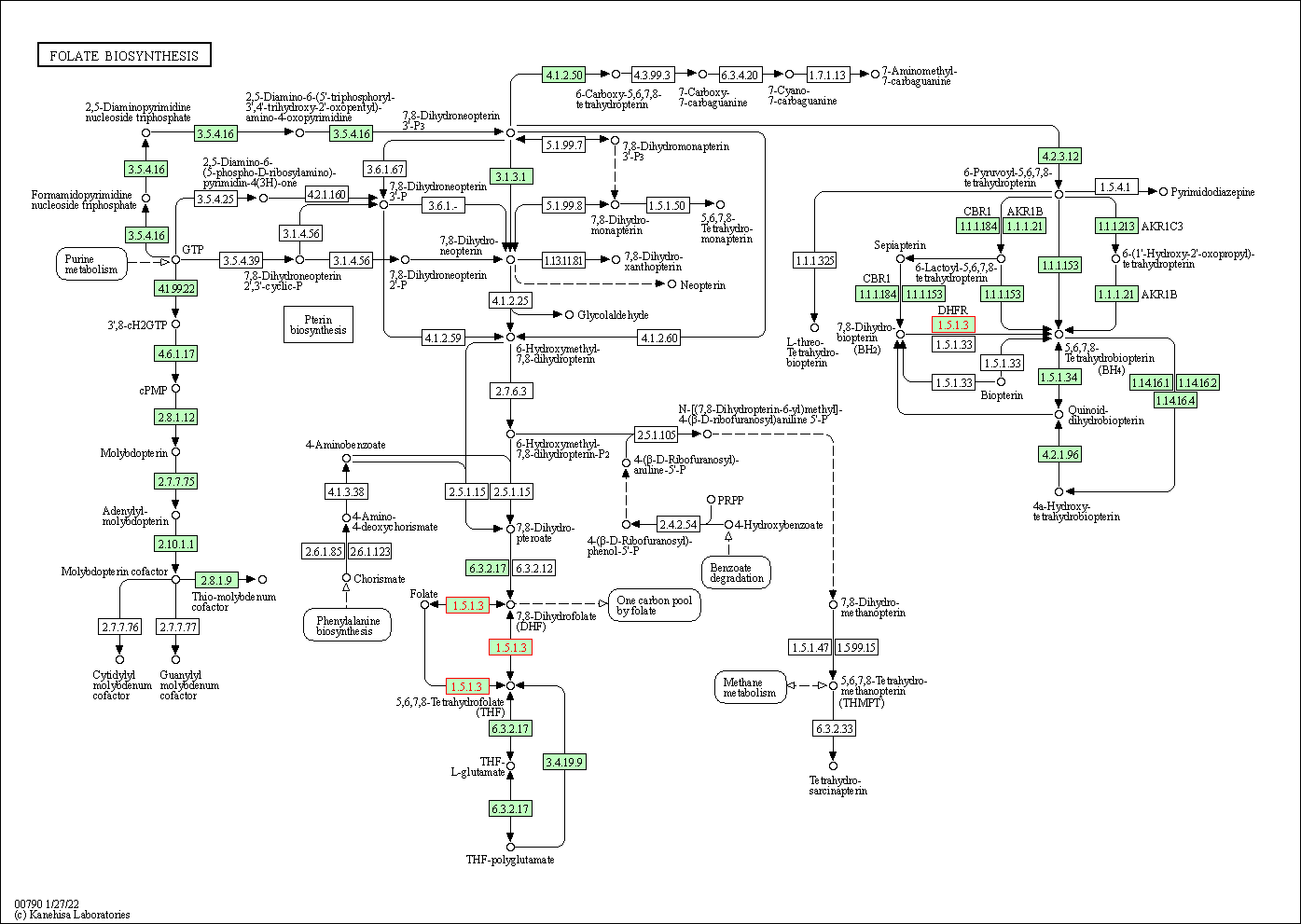
|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Degree | 17 | Degree centrality | 1.83E-03 | Betweenness centrality | 4.62E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.15E-01 | Radiality | 1.38E+01 | Clustering coefficient | 2.79E-01 |
| Neighborhood connectivity | 1.65E+01 | Topological coefficient | 1.12E-01 | Eccentricity | 10 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Drug Resistance Mutation (DRM) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | One carbon pool by folate | |||||
| 2 | Folate biosynthesis | |||||
| 3 | Metabolic pathways | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Tetrahydrofolate biosynthesis | |||||
| 2 | Formyltetrahydroformate biosynthesis | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Folate Metabolism | |||||
| 2 | Pterine Biosynthesis | |||||
| PID Pathway | [+] 1 PID Pathways | + | ||||
| 1 | E2F transcription factor network | |||||
| Reactome | [+] 4 Reactome Pathways | + | ||||
| 1 | E2F mediated regulation of DNA replication | |||||
| 2 | Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation | |||||
| 3 | Metabolism of folate and pterines | |||||
| 4 | G1/S-Specific Transcription | |||||
| WikiPathways | [+] 8 WikiPathways | + | ||||
| 1 | Nucleotide Metabolism | |||||
| 2 | Trans-sulfuration and one carbon metabolism | |||||
| 3 | Retinoblastoma (RB) in Cancer | |||||
| 4 | One Carbon Metabolism | |||||
| 5 | Metabolism of water-soluble vitamins and cofactors | |||||
| 6 | Metabolism of nitric oxide | |||||
| 7 | Folate Metabolism | |||||
| 8 | Fluoropyrimidine Activity | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 3 | Has nature already identified all useful antibacterial targets Curr Opin Microbiol. 2008 Oct;11(5):387-92. | |||||
| REF 4 | How many modes of action should an antibiotic have Curr Opin Pharmacol. 2008 Oct;8(5):564-73. | |||||
| REF 5 | ClinicalTrials.gov (NCT01116141) A Study of CH-4051 in Patients With Rheumatoid Arthritis (RA). U.S. National Institutes of Health. | |||||
| REF 6 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7414). | |||||
| REF 7 | ClinicalTrials.gov (NCT00002914) Piritrexim in Treating Patients With Advanced Cancer of the Urinary Tract. U.S. National Institutes of Health. | |||||
| REF 8 | Clinical pipeline report, company report or official report of Vyera Pharmaceuticals. | |||||
| REF 9 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800006614) | |||||
| REF 10 | Final results of a phase I and pharmacokinetic study of gamma-methylene-10-deazaaminopterin (MDAM) administered intravenously daily for five consec... Cancer Chemother Pharmacol. 2004 May;53(5):370-6. | |||||
| REF 11 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002566) | |||||
| REF 12 | The pharmacokinetics of 1954U89, 1,3-diamino-7-(1-ethylpropyl)-8-methyl-7H-pyrrolo-(3,2-f)quinazoline, in dogs and rats after intravenous and oral administration. Biopharm Drug Dispos. 1997 Jul;18(5):433-42. | |||||
| REF 13 | Biochemical factors in the selectivity of leucovorin rescue: selective inhibition of leucovorin reactivation of dihydrofolate reductase and leucovorin utilization in purine and pyrimidine biosynthesis by methotrexate and dihydrofolate polyglutamates.NCI Monogr.1987;(5):17-26. | |||||
| REF 14 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 15 | CH-1504, a metabolically inert antifolate for the potential treatment of rheumatoid arthritis. IDrugs. 2010 Aug;13(8):559-67. | |||||
| REF 16 | Structural variations of piritrexim, a lipophilic inhibitor of human dihydrofolate reductase: synthesis, antitumor activity and molecular modeling ... Eur J Med Chem. 2004 Dec;39(12):1079-88. | |||||
| REF 17 | Polyglutamylation of the dihydrofolate reductase inhibitor gamma-methylene-10-deazaaminopterin is not essential for antitumor activity. Clin Cancer Res. 1996 Apr;2(4):707-12. | |||||
| REF 18 | Evaluation of the importance of hydrophobic interactions in drug binding to dihydrofolate reductase. J Med Chem. 1988 Jan;31(1):129-37. | |||||
| REF 19 | A Ring-Transformation/Ring-Annulation Strategy for the Synthesis of the DHFR Inhibitor, TNP-351: A Correction. J Org Chem. 1996 Nov 1;61(22):7973-7974. | |||||
| REF 20 | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND A LIPOPHILIC ANTIFOLATE SELECTIVE FOR MYCOBACTERIUM AVIUM DHFR, 6-((2,5- DIETHOXYPHENYL)AMINOMETHYL)-2,4-DIAMINO-5-METHYLPYRIDO(2,3-D) PYRIMIDINE (SRI-8686) | |||||
| REF 21 | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND TRIMETHOPRIM | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

