Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T32578
(Former ID: TTDC00080)
|
|||||
| Target Name |
Interleukin-6 (IL6)
|
|||||
| Synonyms |
Interferon beta-2; IL-6; IFNB2; IFN-beta-2; Hybridoma growth factor; CTL differentiation factor; CDF; BSF-2; B-cell stimulatory factor 2
Click to Show/Hide
|
|||||
| Gene Name |
IL6
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Anemia [ICD-11: 3A00-3A9Z] | |||||
| Function |
It is a potent inducer of the acute phase response. Plays an essential role in the final differentiation of B-cells into Ig-secreting cells Involved in lymphocyte and monocyte differentiation. Acts on B-cells, T-cells, hepatocytes, hematopoietic progenitor cells and cells of the CNS. Required for the generation of T(H)17 cells. Also acts as a myokine. It is discharged into the bloodstream after muscle contraction and acts to increase the breakdown of fats and to improve insulin resistance. It induces myeloma and plasmacytoma growth and induces nerve cells differentiation. Cytokine with a wide variety of biological functions.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine: interleukin
|
|||||
| UniProt ID | ||||||
| Sequence |
MNSFSTSAFGPVAFSLGLLLVLPAAFPAPVPPGEDSKDVAAPHRQPLTSSERIDKQIRYI
LDGISALRKETCNKSNMCESSKEALAENNLNLPKMAEKDGCFQSGFNEETCLVKIITGLL EFEVYLEYLQNRFESSEEQARAVQMSTKVLIQFLQKKAKNLDAITTPDPTTNASLLTKLQ AQNQWLQDMTTHLILRSFKEFLQSSLRALRQM Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A03633 ; BADD_A04013 ; BADD_A05078 ; BADD_A05675 ; BADD_A05823 ; BADD_A05962 ; BADD_A06761 | |||||
| HIT2.0 ID | T33OPP | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Siltuximab | Drug Info | Approved | Anemia | [1], [2], [3] | |
| Clinical Trial Drug(s) | [+] 10 Clinical Trial Drugs | + | ||||
| 1 | Olokizumab | Drug Info | Phase 3 | Rheumatoid arthritis | [6] | |
| 2 | Sirukumab | Drug Info | Phase 3 | Rheumatoid arthritis | [7], [8] | |
| 3 | ALD-518 | Drug Info | Phase 2 | Rheumatoid arthritis | [10] | |
| 4 | CDP-6038 | Drug Info | Phase 2 | Rheumatoid arthritis | [10] | |
| 5 | Clazakizumab | Drug Info | Phase 2 | Crohn disease | [11] | |
| 6 | PF-04236921 | Drug Info | Phase 2 | Systemic lupus erythematosus | [12] | |
| 7 | C326 | Drug Info | Phase 1 | Crohn disease | [14] | |
| 8 | Gerilimzumab | Drug Info | Phase 1 | Rheumatoid arthritis | [6] | |
| 9 | MEDI5117 | Drug Info | Phase 1 | Rheumatoid arthritis | [15] | |
| 10 | OP-R003 | Drug Info | Phase 1 | Haematological malignancy | [16] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 5 Modulator drugs | + | ||||
| 1 | Olokizumab | Drug Info | [17] | |||
| 2 | ALD-518 | Drug Info | [10] | |||
| 3 | CDP-6038 | Drug Info | [10] | |||
| 4 | MEDI5117 | Drug Info | [20] | |||
| 5 | OP-R003 | Drug Info | [21] | |||
| Inhibitor | [+] 4 Inhibitor drugs | + | ||||
| 1 | Sirukumab | Drug Info | [6], [18] | |||
| 2 | Clazakizumab | Drug Info | [6] | |||
| 3 | PF-04236921 | Drug Info | [6] | |||
| 4 | C326 | Drug Info | [19] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Viral protein interaction with cytokine and cytokine receptor | hsa04061 | Affiliated Target |
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Cytosolic DNA-sensing pathway | hsa04623 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| JAK-STAT signaling pathway | hsa04630 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Hematopoietic cell lineage | hsa04640 | Affiliated Target |
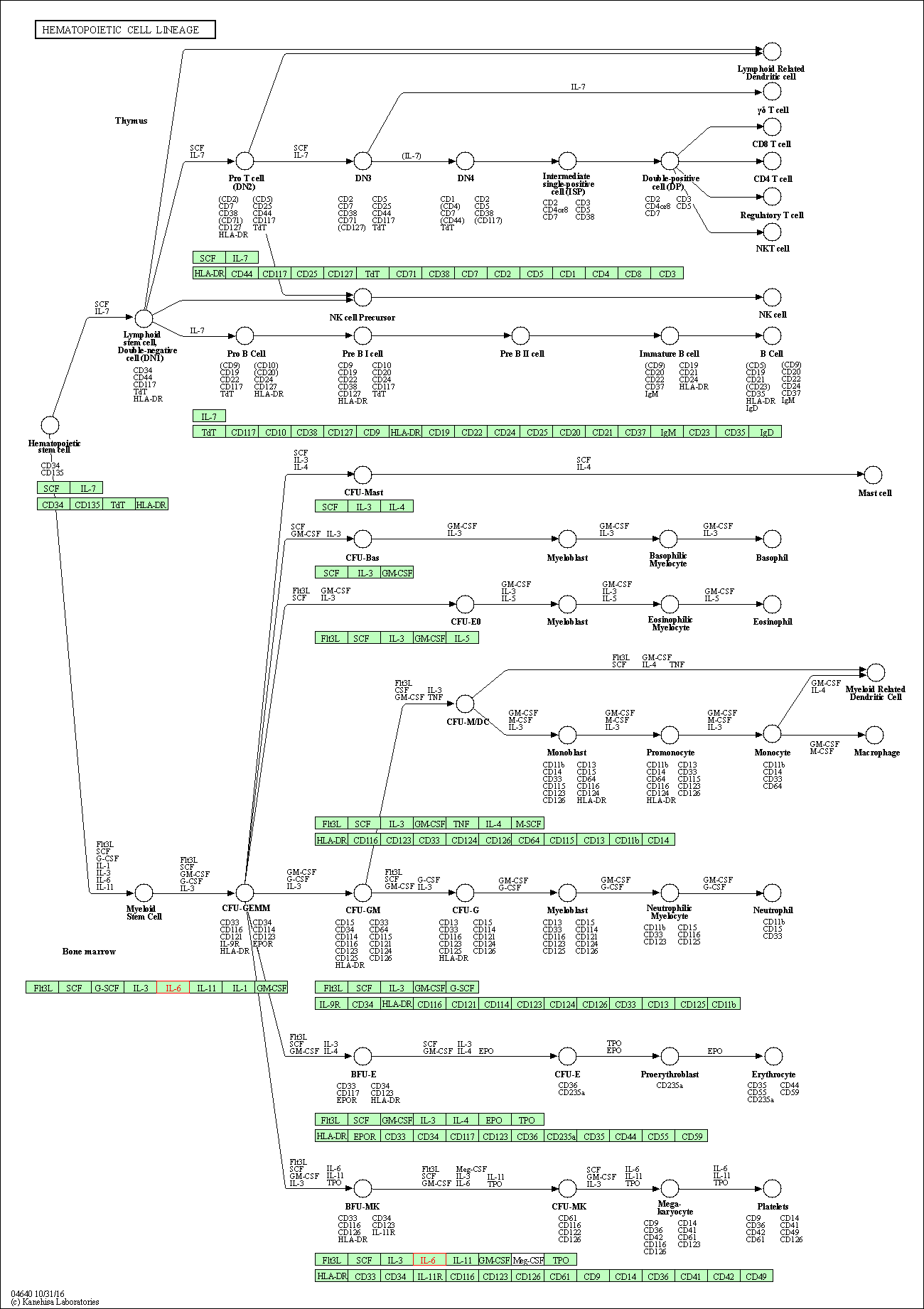
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
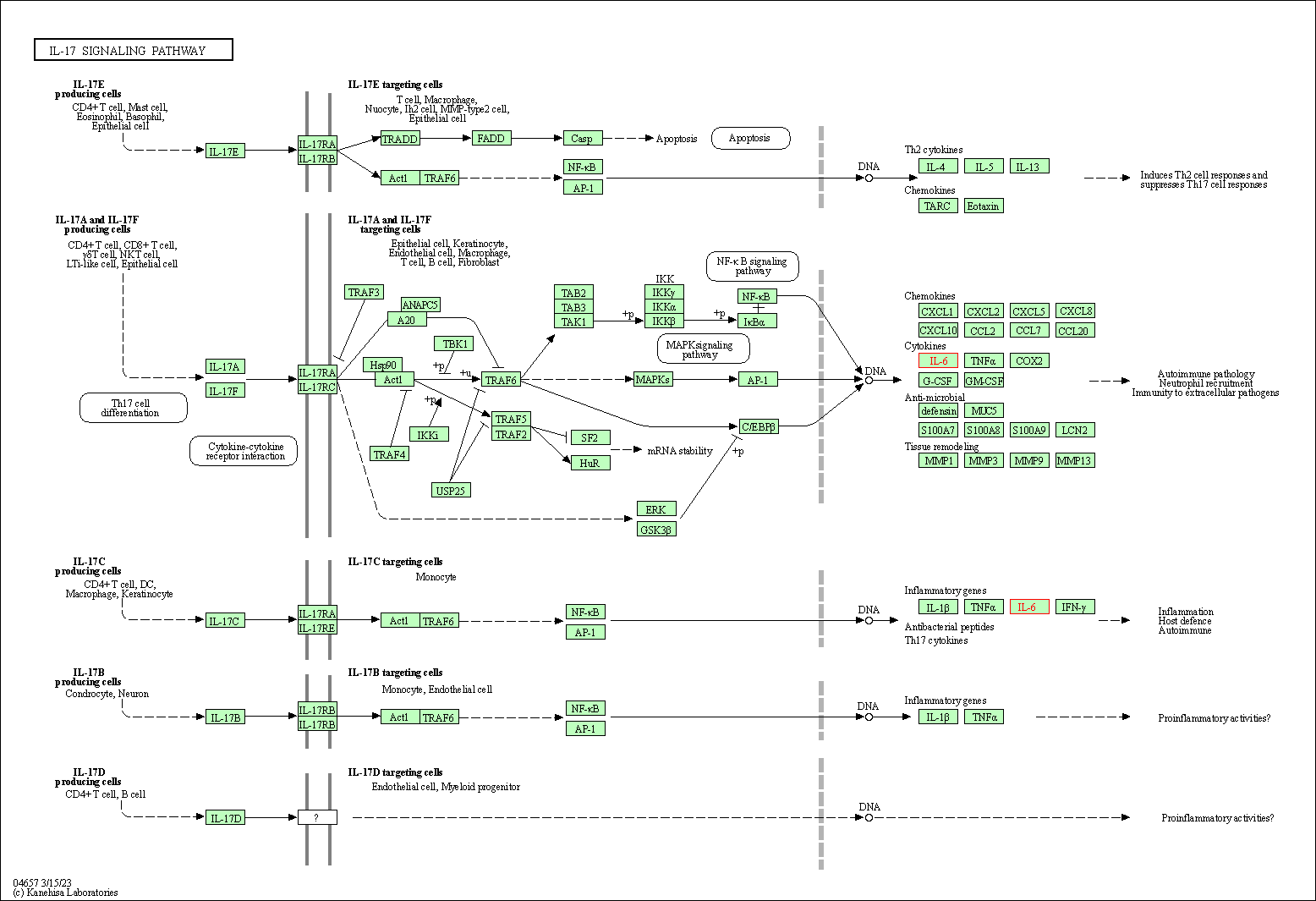
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |
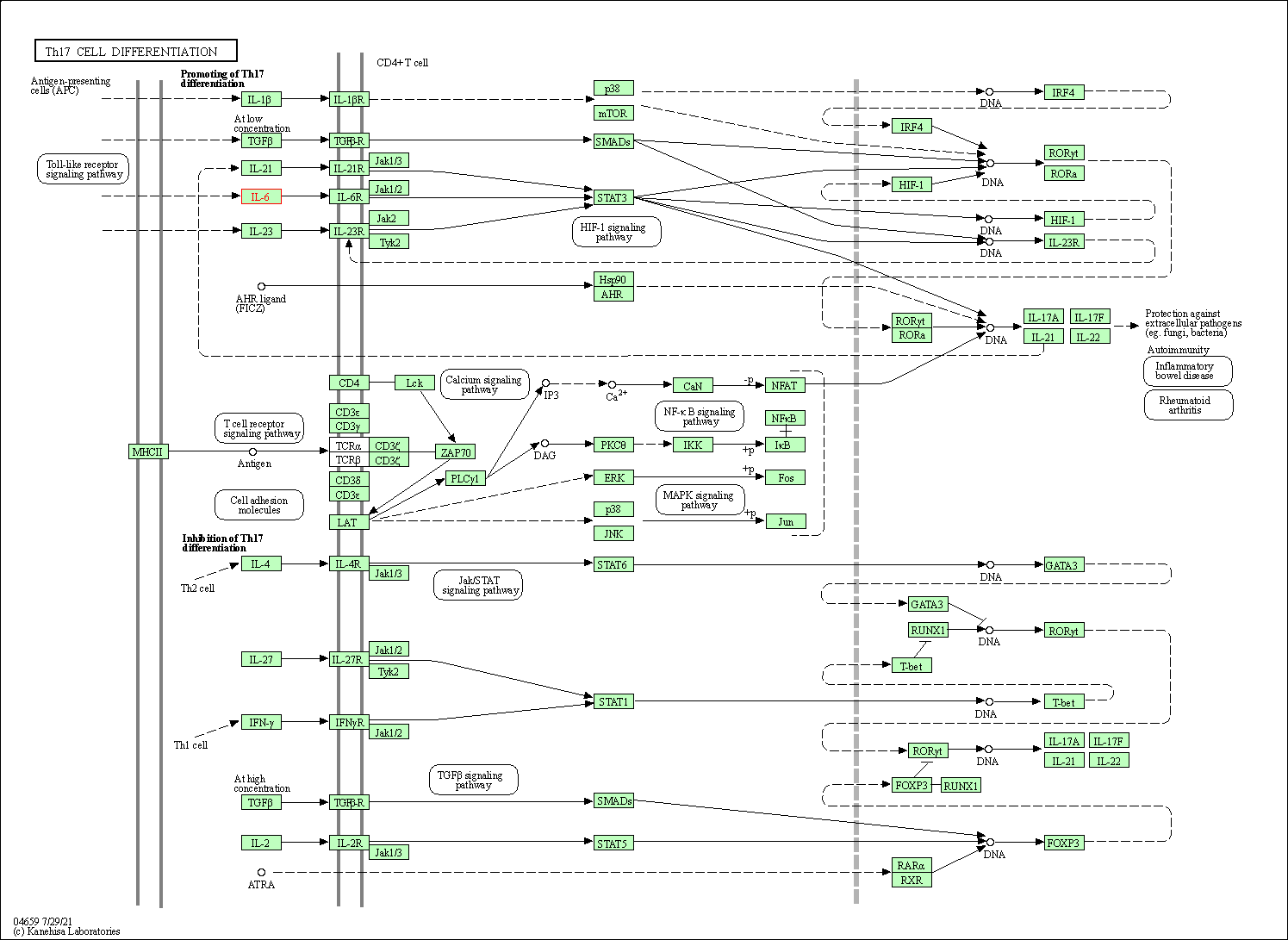
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
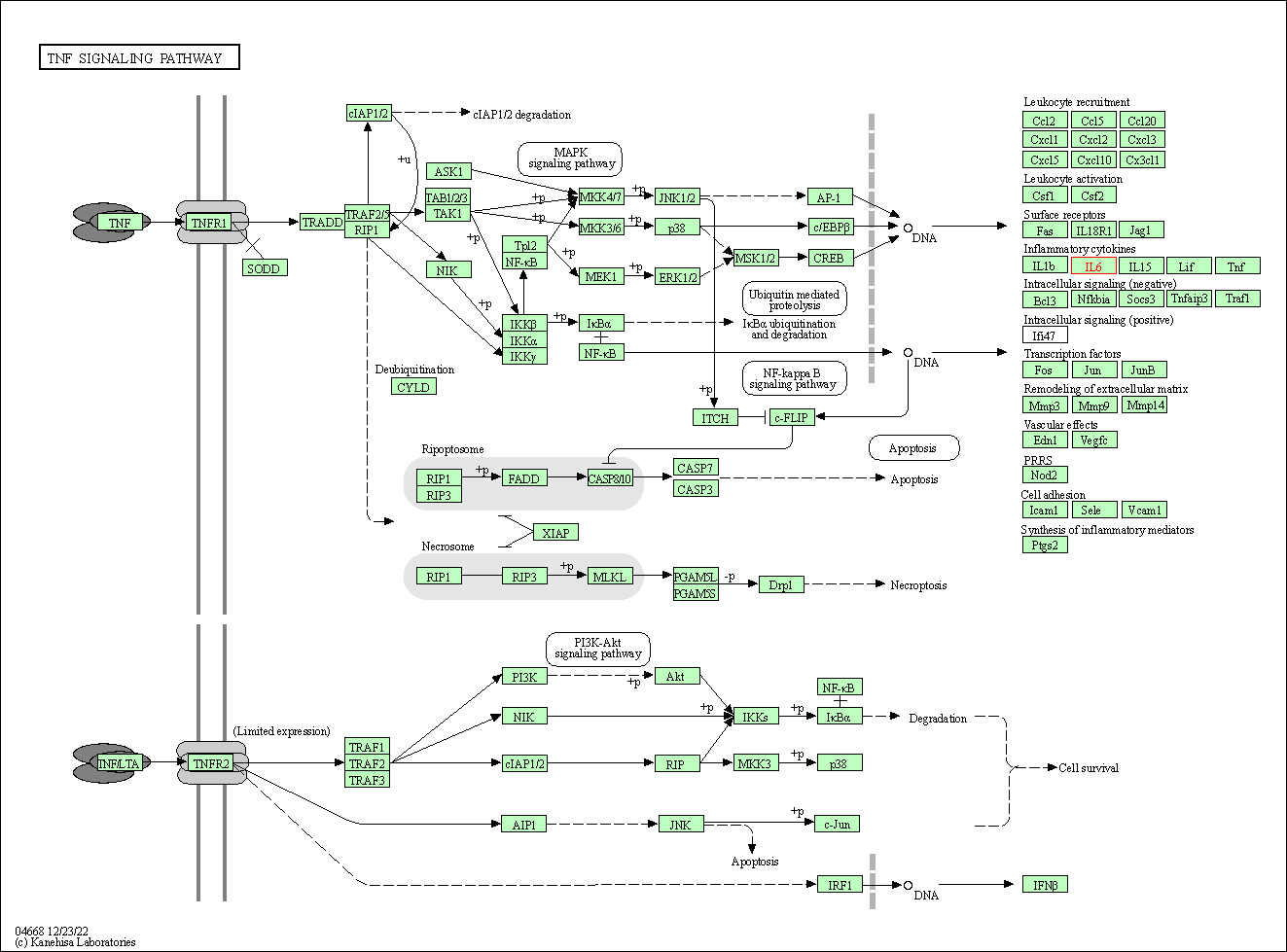
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Intestinal immune network for IgA production | hsa04672 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 62 | Degree centrality | 6.66E-03 | Betweenness centrality | 2.63E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.50E-01 | Radiality | 1.44E+01 | Clustering coefficient | 1.79E-01 |
| Neighborhood connectivity | 3.31E+01 | Topological coefficient | 4.65E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | 2014 FDA drug approvals. Nat Rev Drug Discov. 2015 Feb;14(2):77-81. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7396). | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | ClinicalTrials.gov (NCT05636176) HERMES: Effects of Ziltivekimab Versus Placebo on Morbidity and Mortality in Patients With Heart Failure With Mildly Reduced or Preserved Ejection Fraction and Systemic Inflammation. U.S.National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of Roche | |||||
| REF 6 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 7 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7989). | |||||
| REF 8 | ClinicalTrials.gov (NCT02531633) Efficacy and Safety Study of Sirukumab in Patients With Giant Cell Arteritis. | |||||
| REF 9 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800020380) | |||||
| REF 10 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 11 | ClinicalTrials.gov (NCT02015520) Phase IIB Dose Ranging Study in Subjects With Moderate to Severe Rheumatoid Arthritis. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT01405196) Subcutaneous Treatment In Randomized Subjects To Evaluate Safety And Efficacy In Generalized Lupus Erythematosus. U.S. National Institutes of Health. | |||||
| REF 13 | Clinical pipeline report, company report or official report of Sanofi | |||||
| REF 14 | ClinicalTrials.gov (NCT00353756) Phase 1 Study of Safety and Biological Effects of C326, an Inhibitor of IL-6, in Crohn's Disease. U.S. National Institutes of Health. | |||||
| REF 15 | ClinicalTrials.gov (NCT01559103) Study to Assess the Safety and Tolerability of MEDI5117 in Rheumatoid Arthritis Patients. U.S. National Institutes of Health. | |||||
| REF 16 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800034097) | |||||
| REF 17 | Efficacy and safety of olokizumab in patients with rheumatoid arthritis with an inadequate response to TNF inhibitor therapy: outcomes of a randomised Phase IIb study. Ann Rheum Dis. 2014 September; 73(9): 1607-1615. | |||||
| REF 18 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 19 | The molecular basis of hepcidin-resistant hereditary hemochromatosis. Blood. 2009 Jul 9;114(2):437-43. | |||||
| REF 20 | Whole-molecule antibody engineering: generation of a high-affinity anti-IL-6 antibody with extended pharmacokinetics. J Mol Biol. 2011 Aug 26;411(4):791-807. | |||||
| REF 21 | A high-affinity fully human anti-IL-6 mAb (OP-R003-1, 1339) for the treatment of Multiple Myeloma | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

