Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T36483
(Former ID: TTDC00071)
|
|||||
| Target Name |
Proteinase activated receptor 1 (F2R)
|
|||||
| Synonyms |
Thrombin receptor; Proteinase-activated receptor 1; Protease-activated receptor-1; Protease activated receptor 1; PAR1; PAR-1; Coagulation factor II receptor; CF2R
Click to Show/Hide
|
|||||
| Gene Name |
F2R
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Myocardial infarction [ICD-11: BA41-BA43] | |||||
| Function |
May play a role in platelets activation and in vascular development. High affinity receptor for activated thrombin coupled to G proteins that stimulate phosphoinositide hydrolysis.
Click to Show/Hide
|
|||||
| BioChemical Class |
GPCR rhodopsin
|
|||||
| UniProt ID | ||||||
| Sequence |
MGPRRLLLVAACFSLCGPLLSARTRARRPESKATNATLDPRSFLLRNPNDKYEPFWEDEE
KNESGLTEYRLVSINKSSPLQKQLPAFISEDASGYLTSSWLTLFVPSVYTGVFVVSLPLN IMAIVVFILKMKVKKPAVVYMLHLATADVLFVSVLPFKISYYFSGSDWQFGSELCRFVTA AFYCNMYASILLMTVISIDRFLAVVYPMQSLSWRTLGRASFTCLAIWALAIAGVVPLLLK EQTIQVPGLNITTCHDVLNETLLEGYYAYYFSAFSAVFFFVPLIISTVCYVSIIRCLSSS AVANRSKKSRALFLSAAVFCIFIICFGPTNVLLIAHYSFLSHTSTTEAAYFAYLLCVCVS SISCCIDPLIYYYASSECQRYVYSILCCKESSDPSSYNSSGQLMASKMDTCSSNLNNSIY KKLLT Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T19F7P | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | E5555 | Drug Info | Phase 2 | Acute coronary syndrome | [6], [7] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | RWJ-58259 | Drug Info | Terminated | Artery stenosis | [9] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Antagonist | [+] 3 Antagonist drugs | + | ||||
| 1 | E5555 | Drug Info | [10], [11] | |||
| 2 | RWJ-56110 | Drug Info | [12] | |||
| 3 | SCH-602539 | Drug Info | [14] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | RWJ-58259 | Drug Info | [9] | |||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | SCH-205831 | Drug Info | [13] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Olfactory receptor 11A1 (OR11A1) | 27.184 (28/103) | 1.00E-03 | |
| Olfactory receptor 2AE1 (OR2AE1) | 24.627 (33/134) | 4.00E-03 |
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Calcium signaling pathway | hsa04020 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |
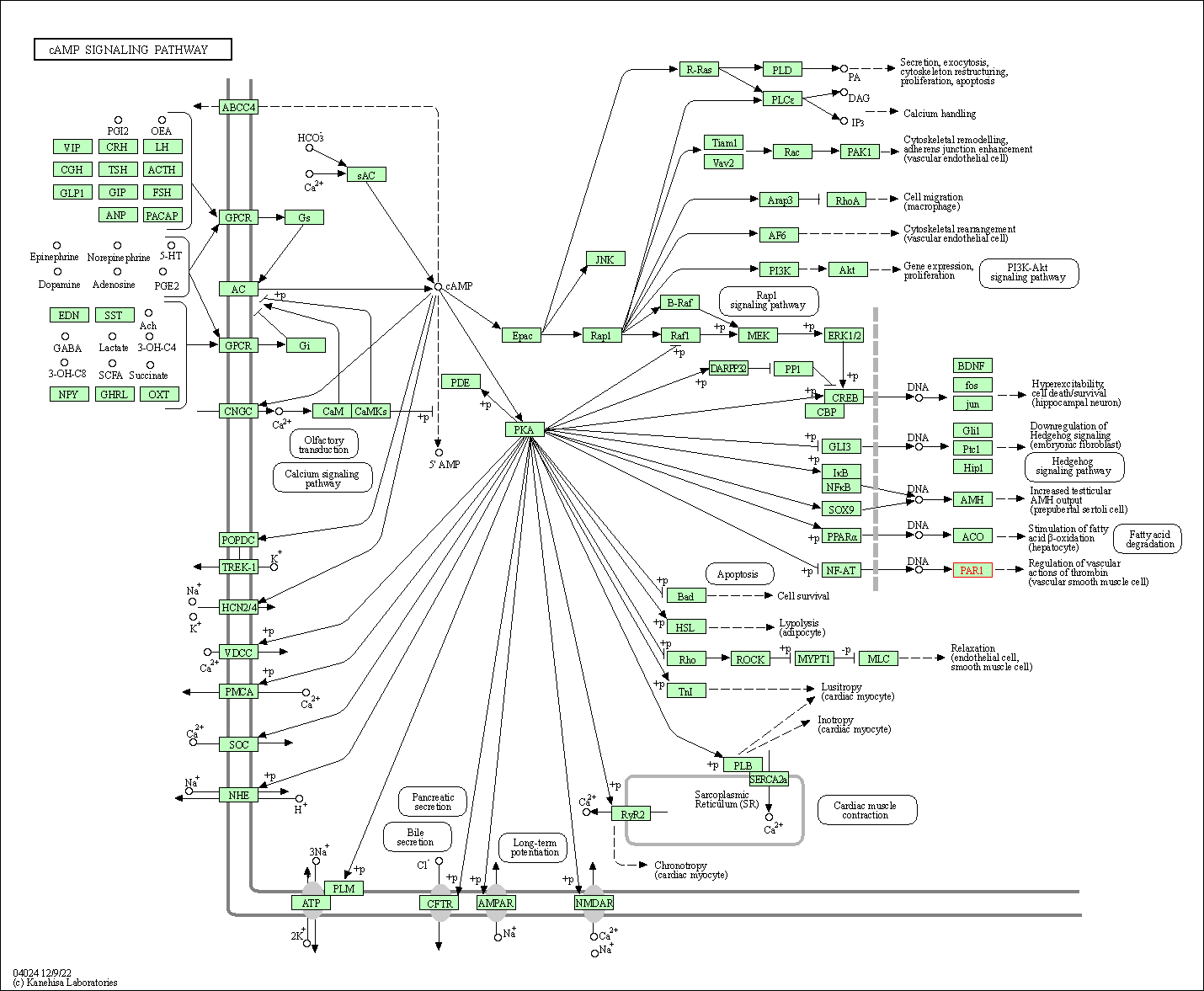
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Neuroactive ligand-receptor interaction | hsa04080 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
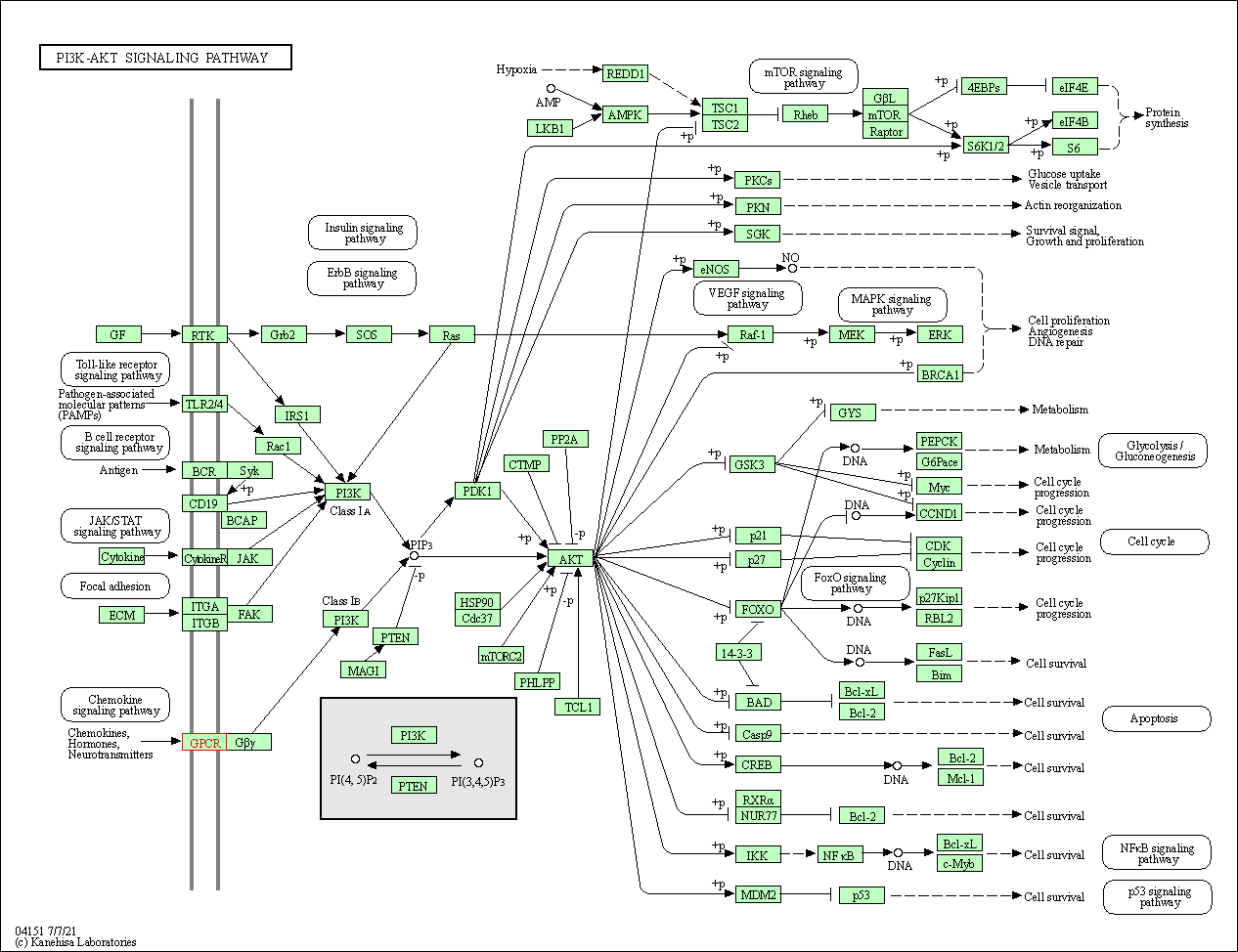
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Complement and coagulation cascades | hsa04610 | Affiliated Target |
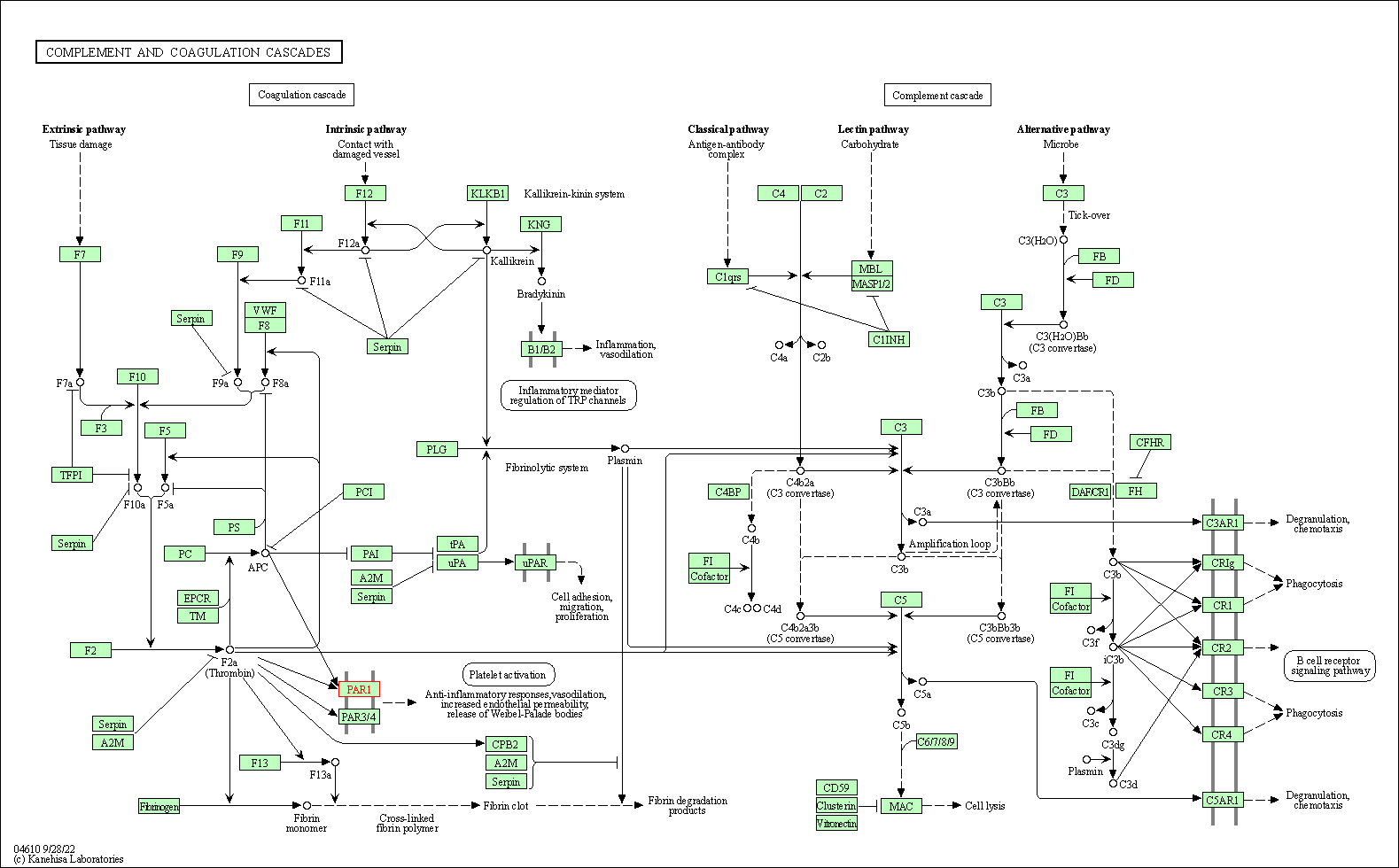
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |
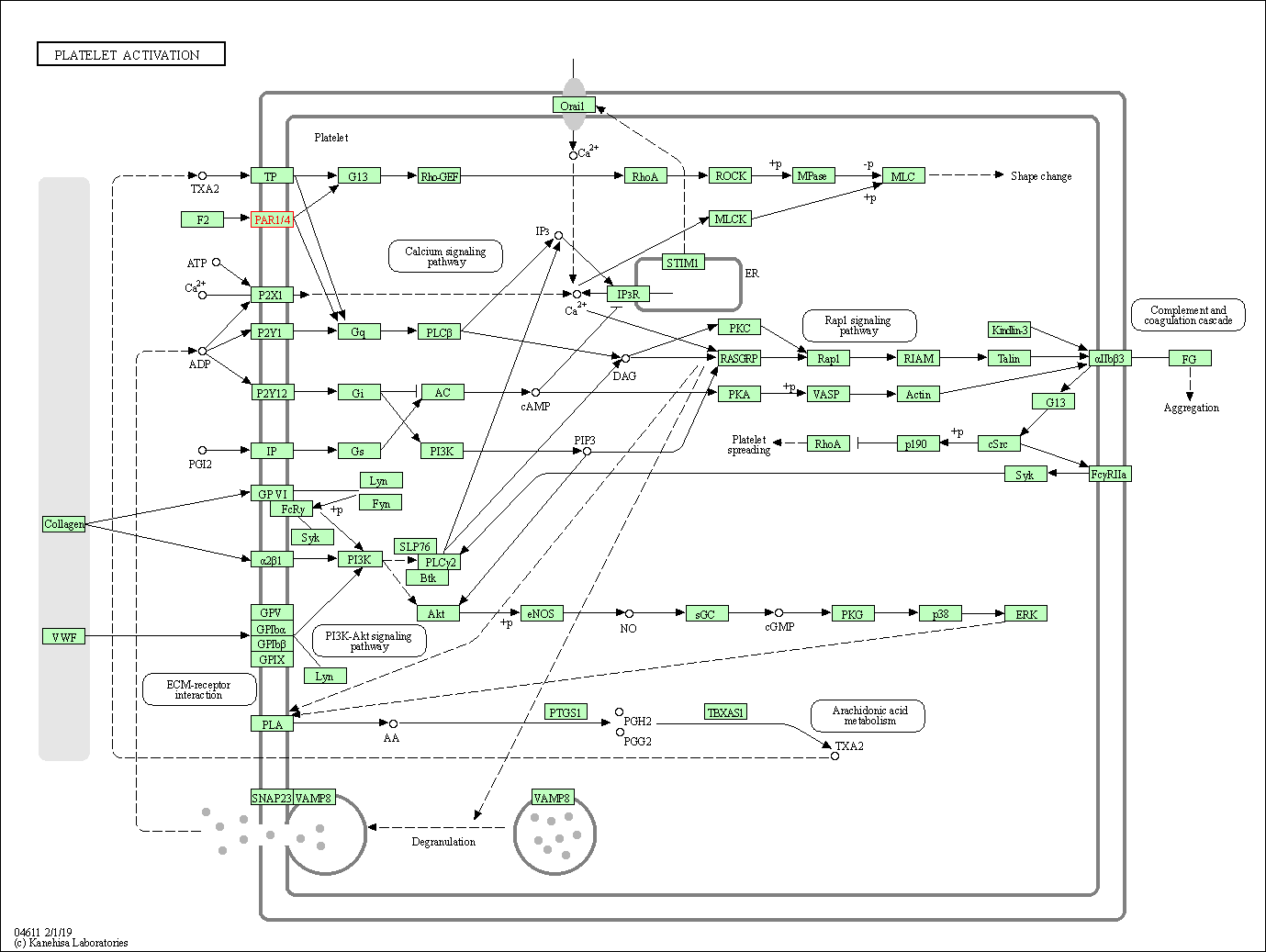
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 5.22E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.09E-01 | Radiality | 1.37E+01 | Clustering coefficient | 1.07E-01 |
| Neighborhood connectivity | 2.11E+01 | Topological coefficient | 1.64E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | In Process Citation. Pharm Unserer Zeit. 2009;38(4):320-8. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4047). | |||||
| REF 3 | 2014 FDA drug approvals. Nat Rev Drug Discov. 2015 Feb;14(2):77-81. | |||||
| REF 4 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 5 | ClinicalTrials.gov (NCT02561000) Safety of PZ-128 in Subjects Undergoing Non-Emergent Percutaneous Coronary Intervention (TRIP-PCI). U.S. National Institutes of Health. | |||||
| REF 6 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4048). | |||||
| REF 7 | ClinicalTrials.gov (NCT00619164) A Double-Blind Study of E5555 in Japanese Patients With Acute Coronary Syndrome. U.S. National Institutes of Health. | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010405) | |||||
| REF 9 | RWJ-58259: a selective antagonist of protease activated receptor-1. Cardiovasc Drug Rev. 2003 Winter;21(4):313-26. | |||||
| REF 10 | Safety and tolerability of atopaxar in the treatment of patients with acute coronary syndromes: the lessons from antagonizing the cellular effects of Thrombin-Acute Coronary Syndromes Trial. Circulation. 2011 May 3;123(17):1843-53. | |||||
| REF 11 | Atopaxar and its effects on markers of platelet activation and inflammation: results from the LANCELOT CAD program. J Thromb Thrombolysis. 2012 Jul;34(1):36-43. | |||||
| REF 12 | Design, synthesis, and biological characterization of a peptide-mimetic antagonist for a tethered-ligand receptor. Proc Natl Acad Sci U S A. 1999 Oct 26;96(22):12257-62. | |||||
| REF 13 | Himbacine derived thrombin receptor (PAR-1) antagonists: SAR of the pyridine ring. Bioorg Med Chem Lett. 2007 Aug 15;17(16):4509-13. | |||||
| REF 14 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 347). | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

