Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T51565
(Former ID: TTDR00257)
|
|||||
| Target Name |
Casein kinase II alpha (CSNK2A1)
|
|||||
| Synonyms |
Protein kinase CK2; Casein kinase II subunit alpha; CK2A1; CK II alpha; CK II
Click to Show/Hide
|
|||||
| Gene Name |
CSNK2A1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Coronavirus infection [ICD-11: 1D92] | |||||
| 2 | Liver cancer [ICD-11: 2C12] | |||||
| 3 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Regulates numerous cellular processes, such as cell cycle progression, apoptosis and transcription, as well as viral infection. May act as a regulatory node which integrates and coordinates numerous signals leading to an appropriate cellular response. During mitosis, functions as a component of the p53/TP53-dependent spindle assembly checkpoint (SAC) that maintains cyclin-B-CDK1 activity and G2 arrest in response to spindle damage. Also required for p53/TP53-mediated apoptosis, phosphorylating 'Ser-392' of p53/TP53 following UV irradiation. Can also negatively regulate apoptosis. Phosphorylates the caspases CASP9 and CASP2 and the apoptotic regulator NOL3. Phosphorylation protects CASP9 from cleavage and activation by CASP8, and inhibits the dimerization of CASP2 and activation of CASP8. Regulates transcription by direct phosphorylation of RNA polymerases I, II, III and IV. Also phosphorylates and regulates numerous transcription factors including NF-kappa-B, STAT1, CREB1, IRF1, IRF2, ATF1, SRF, MAX, JUN, FOS, MYC and MYB. Phosphorylates Hsp90 and its co-chaperones FKBP4 and CDC37, which is essential for chaperone function. Regulates Wnt signaling by phosphorylating CTNNB1 and the transcription factor LEF1. Acts as an ectokinase that phosphorylates several extracellular proteins. During viral infection, phosphorylates various proteins involved in the viral life cycles of EBV, HSV, HBV, HCV, HIV, CMV and HPV. Phosphorylates PML at 'Ser-565' and primes it for ubiquitin-mediated degradation. Plays an important role in the circadian clock function by phosphorylating ARNTL/BMAL1 at 'Ser-90' which is pivotal for its interaction with CLOCK and which controls CLOCK nuclear entry. Phosphorylates CCAR2 at 'Thr-454' in gastric carcinoma tissue. Catalytic subunit of a constitutively active serine/threonine-protein kinase complex that phosphorylates a large number of substrates containing acidic residues C-terminal to the phosphorylated serine or threonine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MSGPVPSRARVYTDVNTHRPREYWDYESHVVEWGNQDDYQLVRKLGRGKYSEVFEAINIT
NNEKVVVKILKPVKKKKIKREIKILENLRGGPNIITLADIVKDPVSRTPALVFEHVNNTD FKQLYQTLTDYDIRFYMYEILKALDYCHSMGIMHRDVKPHNVMIDHEHRKLRLIDWGLAE FYHPGQEYNVRVASRYFKGPELLVDYQMYDYSLDMWSLGCMLASMIFRKEPFFHGHDNYD QLVRIAKVLGTEDLYDYIDKYNIELDPRFNDILGRHSRKRWERFVHSENQHLVSPEALDF LDKLLRYDHQSRLTAREAMEHPYFYTVVKDQARMGSSSMPGGSTPVSSANMMSGISSVPT PSPLGPLAGSPVIAAANPLGMPVPAAAGAQQ Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T40GZM | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | CX-4945 | Drug Info | Phase 2 | Coronavirus infection | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 14 Inhibitor drugs | + | ||||
| 1 | CX-4945 | Drug Info | [1] | |||
| 2 | BALANOL | Drug Info | [3] | |||
| 3 | 4,5,6,7-tetrabromo-1H-benzimidazole | Drug Info | [4] | |||
| 4 | 4,5,6,7-tetrabromo-1H-benzo[d][1,2,3]triazole | Drug Info | [5] | |||
| 5 | 4-[(3,5-diamino-1H-pyrazol-4-yl)diazenyl]phenol | Drug Info | [6] | |||
| 6 | 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole | Drug Info | [5] | |||
| 7 | AdoC(Dpr)2AlaArg6 | Drug Info | [7] | |||
| 8 | APIGENIN | Drug Info | [5] | |||
| 9 | CGP-029482 | Drug Info | [5] | |||
| 10 | CX-5011 | Drug Info | [8] | |||
| 11 | CX-7000 | Drug Info | [8] | |||
| 12 | ELLAGIC ACID | Drug Info | [5] | |||
| 13 | PMID22115617C2c | Drug Info | [9] | |||
| 14 | PMID24900749C1a | Drug Info | [10] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Vitamin B3 | Ligand Info | |||||
| Structure Description | Crystal structure of human CK2a | PDB:3WAR | ||||
| Method | X-ray diffraction | Resolution | 1.04 Å | Mutation | No | [11] |
| PDB Sequence |
SGPVPSRARV
11 YTDVNTHRPR21 EYWDYESHVV31 EWGNQDDYQL41 VRKLGRGKYS51 EVFEAINITN 61 NEKVVVKILK71 PVKKKKIKRE81 IKILENLRGG91 PNIITLADIV101 KDPVSRTPAL 111 VFEHVNNTDF121 KQLYQTLTDY131 DIRFYMYEIL141 KALDYCHSMG151 IMHRDVKPHN 161 VMIDHEHRKL171 RLIDWGLAEF181 YHPGQEYNVR191 VASRYFKGPE201 LLVDYQMYDY 211 SLDMWSLGCM221 LASMIFRKEP231 FFHGHDNYDQ241 LVRIAKVLGT251 EDLYDYIDKY 261 NIELDPRFND271 ILGRHSRKRW281 ERFVHSENQH291 LVSPEALDFL301 DKLLRYDHQS 311 RLTAREAMEH321 PYFYTVVKDQ331 ARMG
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Structure of Compound 17 bound to CK2alpha | PDB:8AEC | ||||
| Method | X-ray diffraction | Resolution | 1.09 Å | Mutation | Yes | [12] |
| PDB Sequence |
GPVPSRARVY
12 TDVNTHRPSE22 YWDYESHVVE32 WGNQDDYQLV42 RKLGRGKYSE52 VFEAINITNN 62 EKVVVKILKP72 VAAAKIKREI82 KILENLRGGP92 NIITLADIVK102 DPVSRTPALV 112 FEHVNNTDFK122 QLYQTLTDYD132 IRFYMYEILK142 ALDYCHSMGI152 MHRDVKPHNV 162 MIDHEHRKLR172 LIDWGLAEFY182 HPGQEYNVRV192 ASRYFKGPEL202 LVDYQMYDYS 212 LDMWSLGCML222 ASMIFRKEPF232 FHGHDNYDQL242 VRIAKVLGTE252 DLYDYIDKYN 262 IELDPRFNDI272 LGRHSRKRWE282 RFVHSENQHL292 VSPEALDFLD302 KLLRYDHQSR 312 LTAREAMEHP322 YFYTVVK
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ribosome biogenesis in eukaryotes | hsa03008 | Affiliated Target |
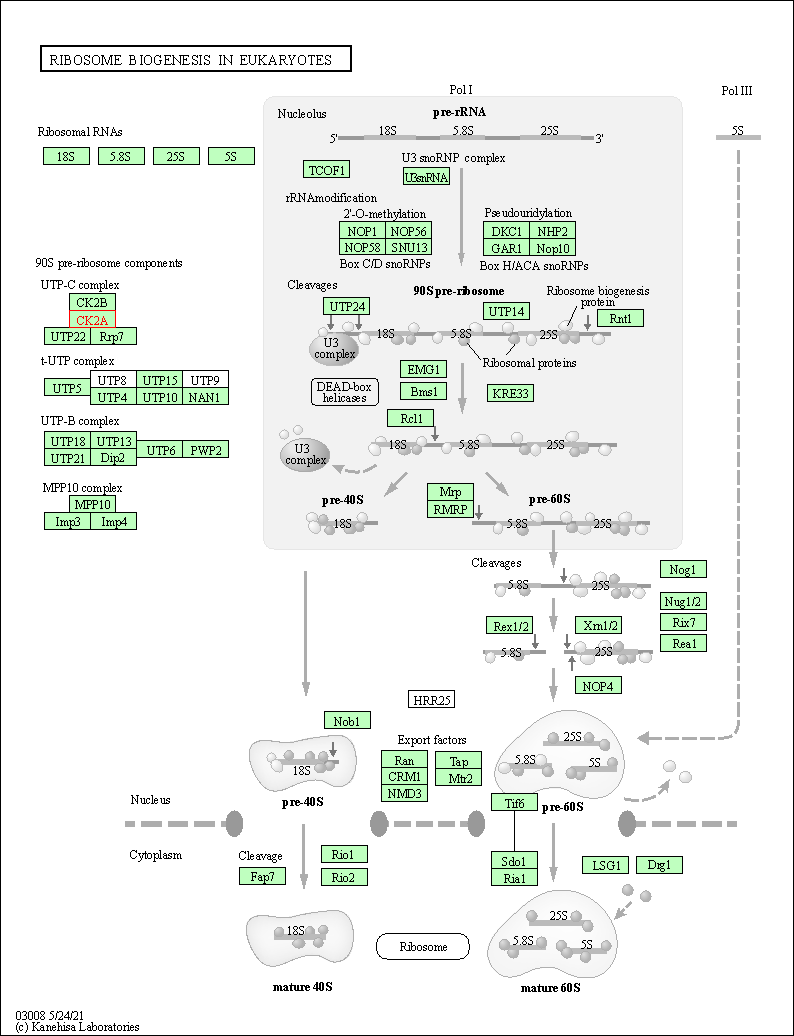
|
| Class: Genetic Information Processing => Translation | Pathway Hierarchy | ||
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
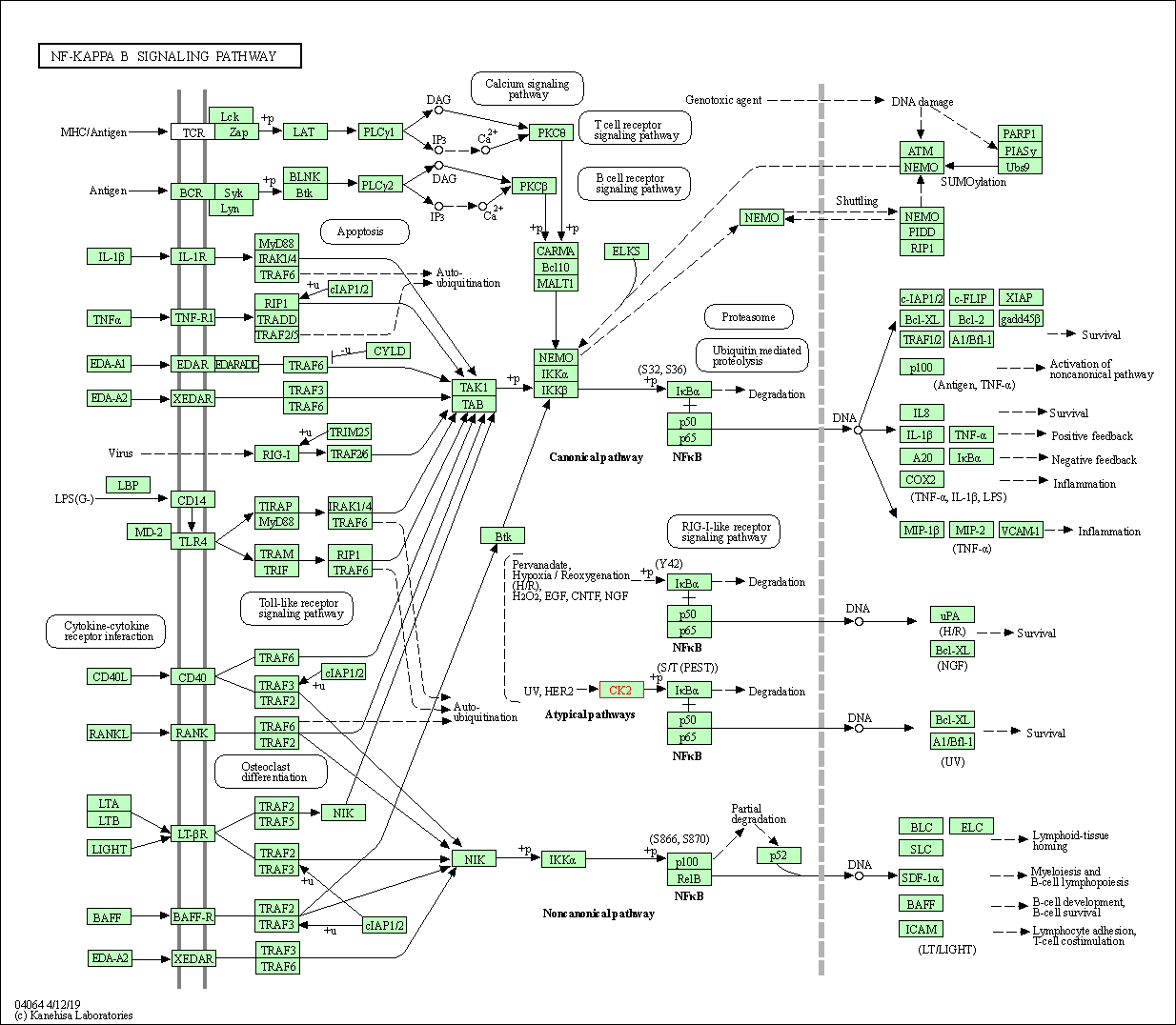
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Mitophagy - animal | hsa04137 | Affiliated Target |
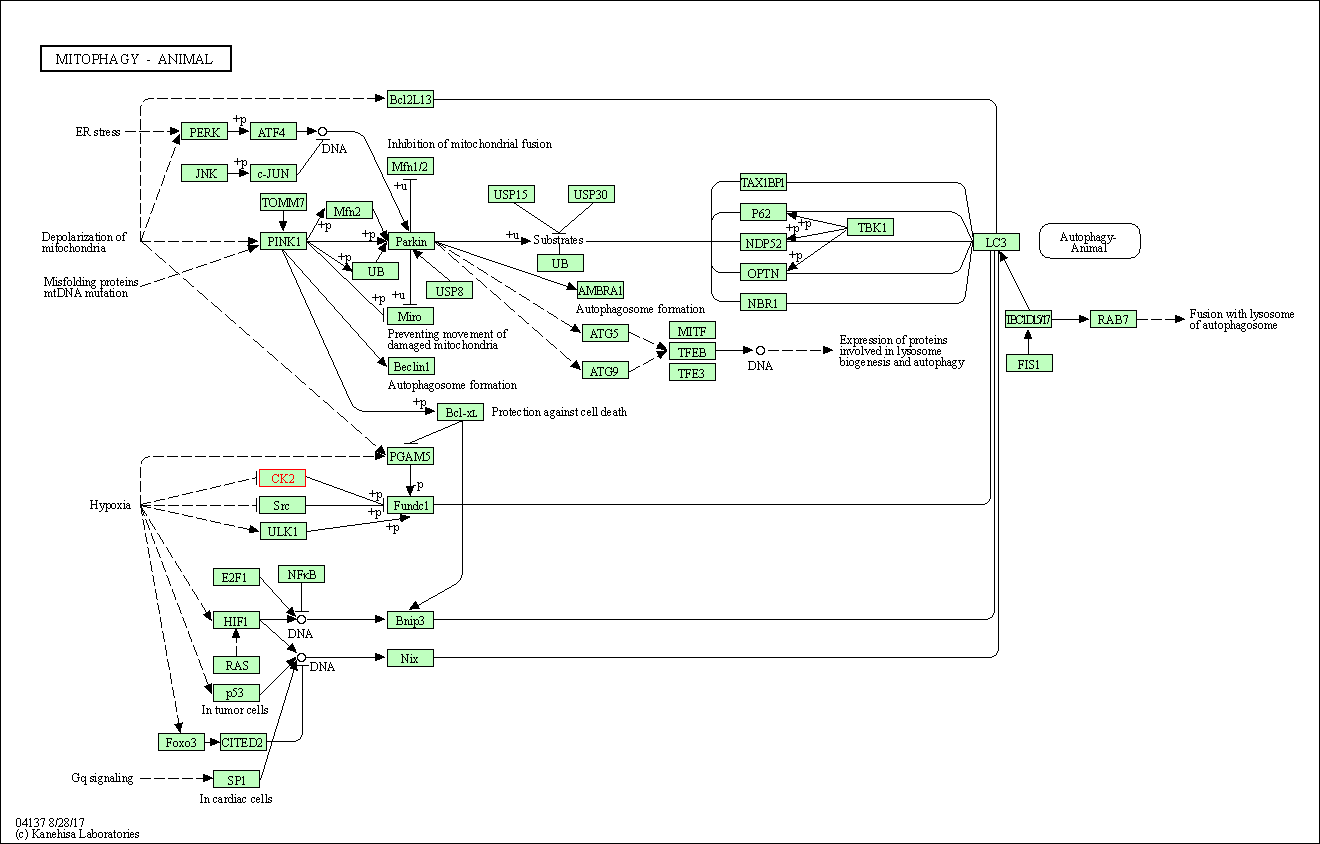
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Wnt signaling pathway | hsa04310 | Affiliated Target |
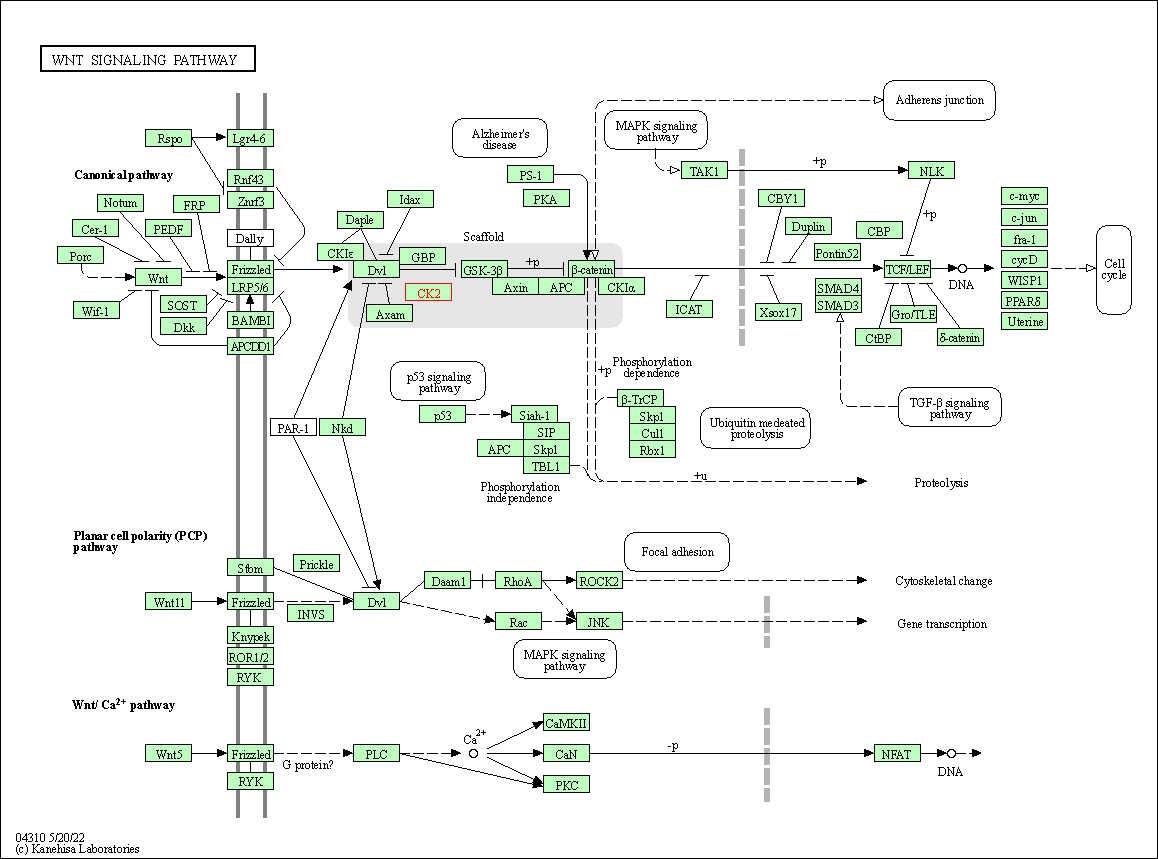
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Adherens junction | hsa04520 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Degree | 15 | Degree centrality | 1.61E-03 | Betweenness centrality | 7.93E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.50E-01 | Radiality | 1.43E+01 | Clustering coefficient | 1.71E-01 |
| Neighborhood connectivity | 5.79E+01 | Topological coefficient | 9.65E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT04668209) Silmitasertib (CX-4945) in Patients With Severe Coronavirus Disease 2019 (COVID-19) (CX4945). U.S. National Institutes of Health. | |||||
| REF 3 | Synthesis and protein kinase C inhibitory activities of acyclic balanol analogs that are highly selective for protein kinase C over protein kinase A. J Med Chem. 1996 Dec 20;39(26):5215-27. | |||||
| REF 4 | Synthesis of new analogs of benzotriazole, benzimidazole and phthalimide--potential inhibitors of human protein kinase CK2. Bioorg Med Chem. 2009 Feb 15;17(4):1573-8. | |||||
| REF 5 | Structural insight into human CK2alpha in complex with the potent inhibitor ellagic acid. Bioorg Med Chem Lett. 2009 Jun 1;19(11):2920-3. | |||||
| REF 6 | 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J Med Chem. 2006 Nov 2;49(22):6500-9. | |||||
| REF 7 | Adenosine-5'-carboxylic acid peptidyl derivatives as inhibitors of protein kinases. Bioorg Med Chem Lett. 1999 May 17;9(10):1447-52. | |||||
| REF 8 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1549). | |||||
| REF 9 | CK2alpha and CK2alpha' subunits differ in their sensitivity to 4,5,6,7-tetrabromo- and 4,5,6,7-tetraiodo-1H-benzimidazole derivatives. Eur J Med Chem. 2012 Jan;47(1):345-50. | |||||
| REF 10 | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo. ACS Med Chem Lett. 2013 Jul 3;4(8):800-5. | |||||
| REF 11 | Crystal structure of human CK2Alpha at 1.06 ? resolution. J Synchrotron Radiat. 2013 Nov;20(Pt 6):974-9. | |||||
| REF 12 | A fragment-based approach leading to the discovery of inhibitors of CK2Alpha with a novel mechanism of action. RSC Med Chem. 2022 Sep 16;13(11):1420-1426. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

