Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T56510
(Former ID: TTDC00168)
|
|||||
| Target Name |
Apoptosis regulator Bcl-xL (BCL-xL)
|
|||||
| Synonyms |
Bcl2like protein 1; Bcl2L1; Bcl2-L-1; Bcl-XL; Bcl-2-like protein 1; BCLX; BCL2L; Apoptosis regulator Bcl-X
Click to Show/Hide
|
|||||
| Gene Name |
BCL2L1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Myeloproliferative neoplasm [ICD-11: 2A20] | |||||
| Function |
Inhibits activation of caspases. Appears to regulate cell death by blocking the voltage-dependent anion channel (VDAC) by binding to it and preventing the release of the caspase activator, CYC1, from the mitochondrial membrane. Also acts as a regulator of G2 checkpoint and progression to cytokinesis during mitosis. Potent inhibitor of cell death.
Click to Show/Hide
|
|||||
| BioChemical Class |
B-cell lymphoma Bcl-2
|
|||||
| UniProt ID | ||||||
| Sequence |
MSQSNRELVVDFLSYKLSQKGYSWSQFSDVEENRTEAPEGTESEMETPSAINGNPSWHLA
DSPAVNGATGHSSSLDAREVIPMAAVKQALREAGDEFELRYRRAFSDLTSQLHITPGTAY QSFEQVVNELFRDGVNWGRIVAFFSFGGALCVESVDKEMQVLVSRIAAWMATYLNDHLEP WIQENGGWDTFVELYGNNAAAESRKGQERFNRWFLTGMTVAGVVLLGSLFSRK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T74I49 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | ABT-263 | Drug Info | Phase 3 | Myelofibrosis | [2] | |
| 2 | Obatoclax | Drug Info | Phase 2 | Solid tumour/cancer | [1] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 11 Inhibitor drugs | + | ||||
| 1 | ABT-263 | Drug Info | [5], [6], [7] | |||
| 2 | Obatoclax | Drug Info | [1] | |||
| 3 | Indole-based analog 2 | Drug Info | [8] | |||
| 4 | Indole-based analog 3 | Drug Info | [8] | |||
| 5 | PMID27744724-Compound-10 | Drug Info | [8] | |||
| 6 | PMID27744724-Compound-18 | Drug Info | [8] | |||
| 7 | PMID27744724-Compound-21 | Drug Info | [8] | |||
| 8 | PMID27744724-Compound-26 | Drug Info | [8] | |||
| 9 | PMID27744724-Compound-27 | Drug Info | [8] | |||
| 10 | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID | Drug Info | [9] | |||
| 11 | WEHI-0103122 | Drug Info | [10] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | E-003 | Drug Info | [10] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Venetoclax | Ligand Info | |||||
| Structure Description | crystal structure of BCL-2 with venetoclax | PDB:6O0K | ||||
| Method | X-ray diffraction | Resolution | 1.62 Å | Mutation | No | [11] |
| PDB Sequence |
YDNREIVMKY
18 IHYKLSQRGY28 EWDAGDDSEV92 VHLTLRQAGD102 DFSRRYRRDF112 AEMSSQLHLT 122 PFTARGRFAT132 VVEELFRDGV142 NWGRIVAFFE152 FGGVMCVESV162 NREMSPLVDN 172 IALWMTEYLN182 RHLHTWIQDN192 GGWDAFVELY202 G
|
|||||
|
|
GLN99
4.770
ALA100
3.123
ASP103
2.772
PHE104
3.735
ARG107
3.306
TYR108
3.688
ASP111
3.665
PHE112
3.777
MET115
3.612
VAL133
4.143
GLU136
3.833
LEU137
3.658
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID | Ligand Info | |||||
| Structure Description | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | PDB:1YSG | ||||
| Method | Solution NMR | Resolution | N.A. | Mutation | No | [12] |
| PDB Sequence |
MSMAMSQSNR
10 ELVVDFLSYK20 LSQKGYSWSQ30 FSDVEENRTE40 APEGTESEAV90 KQALREAGDE 100 FELRYRRAFS110 DLTSQLHITP120 GTAYQSFEQV130 VNELFRDGVN140 WGRIVAFFSF 150 GGALCVESVD160 KEMQVLVSRI170 AAWMATYLND180 HLEPWIQENG190 GWDTFVELYG 200 NNAAAESRKG210 QERLEHHHHH220 H
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Apoptosis facilitator Bcl-2-like protein 14 (BCL2L14) | 24.800 (31/125) | 1.06E-05 | |
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ras signaling pathway | hsa04014 | Affiliated Target |
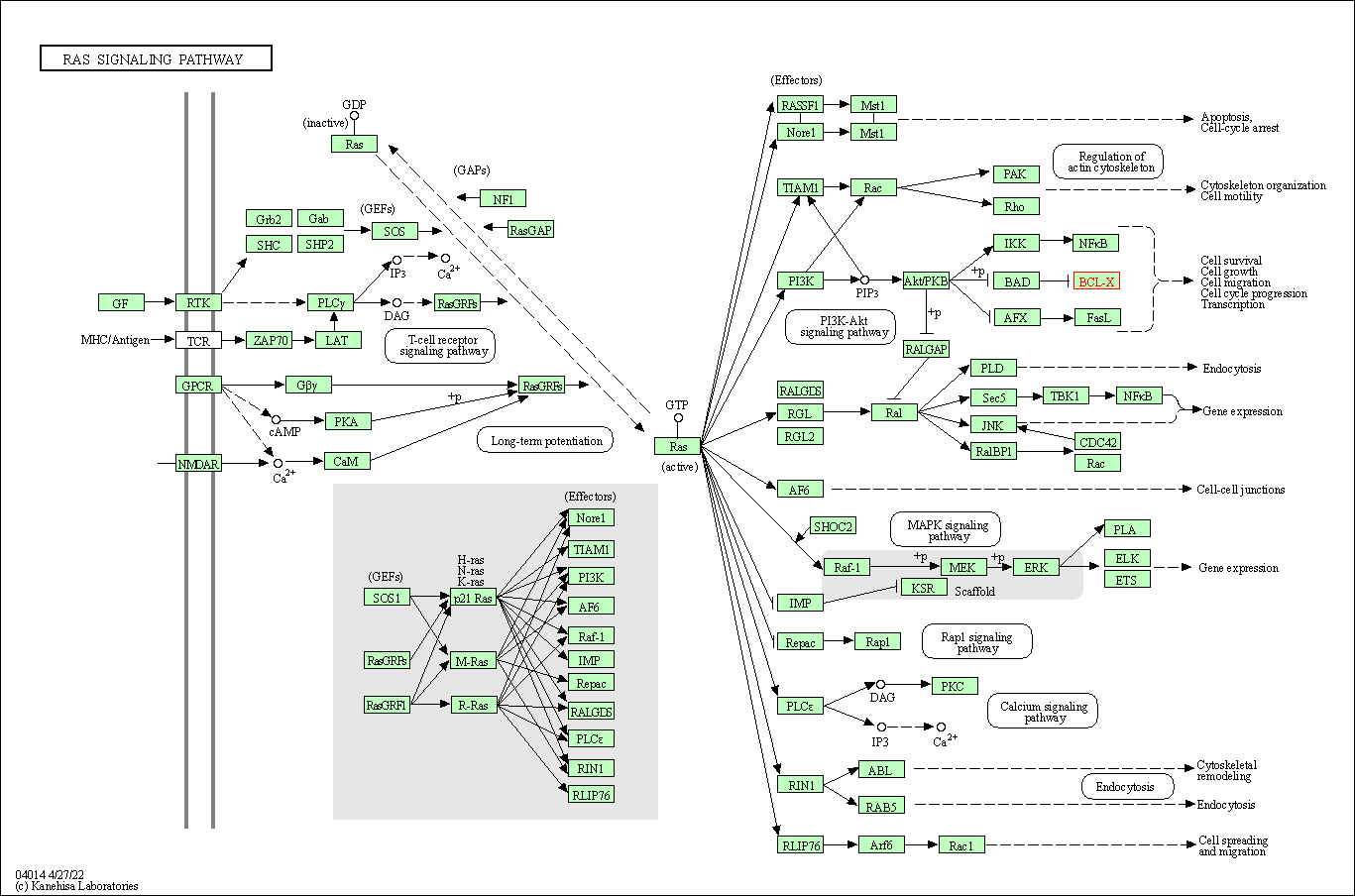
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
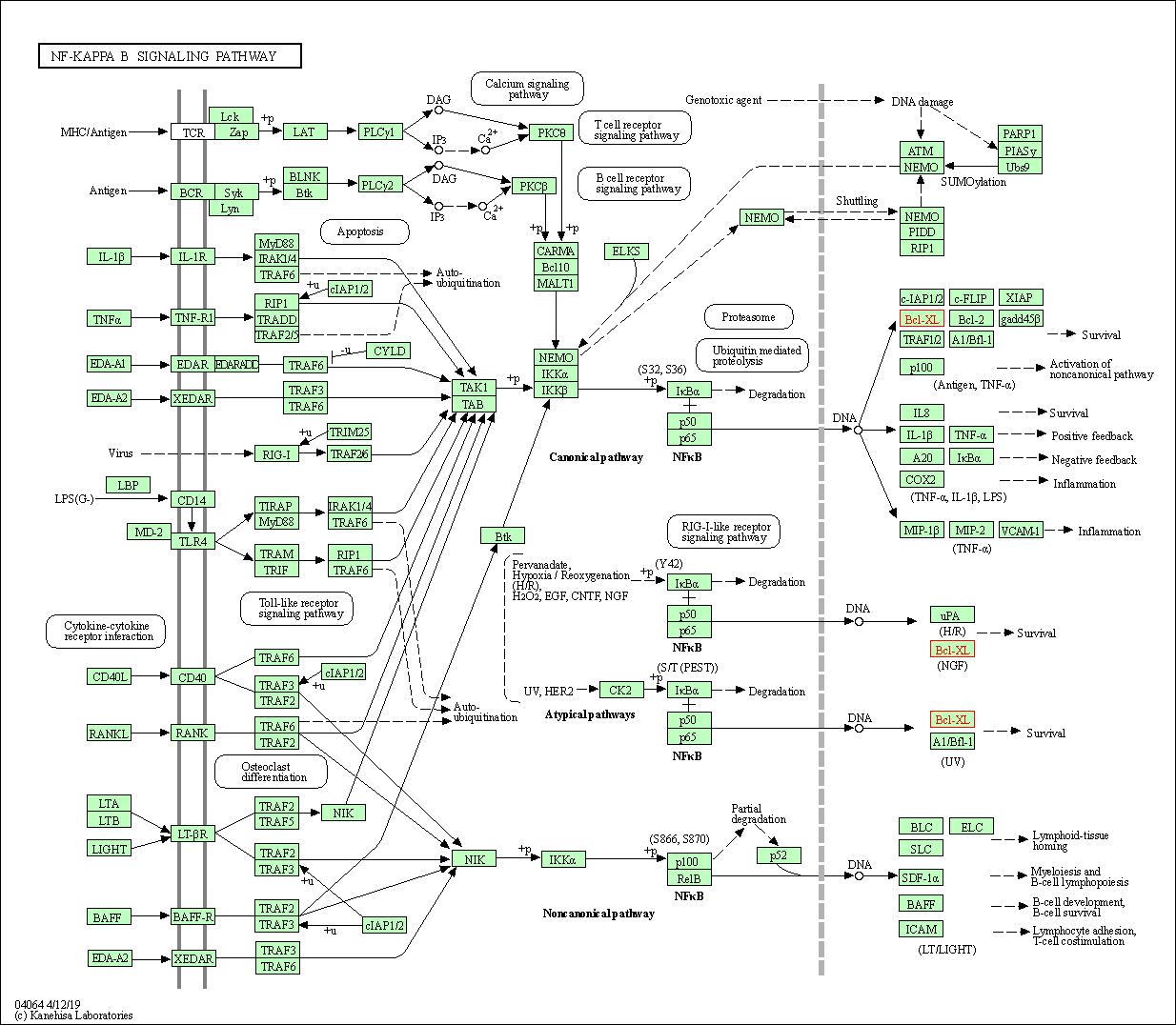
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| p53 signaling pathway | hsa04115 | Affiliated Target |
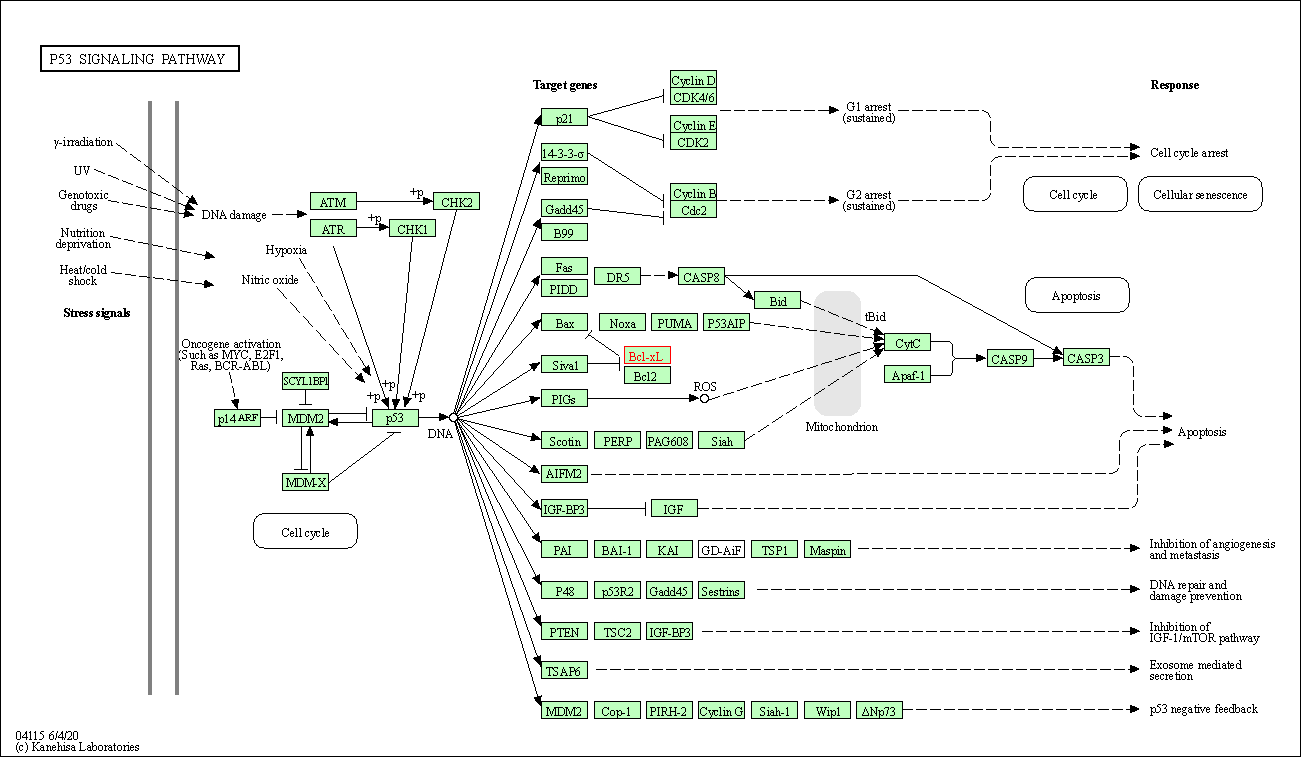
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Mitophagy - animal | hsa04137 | Affiliated Target |
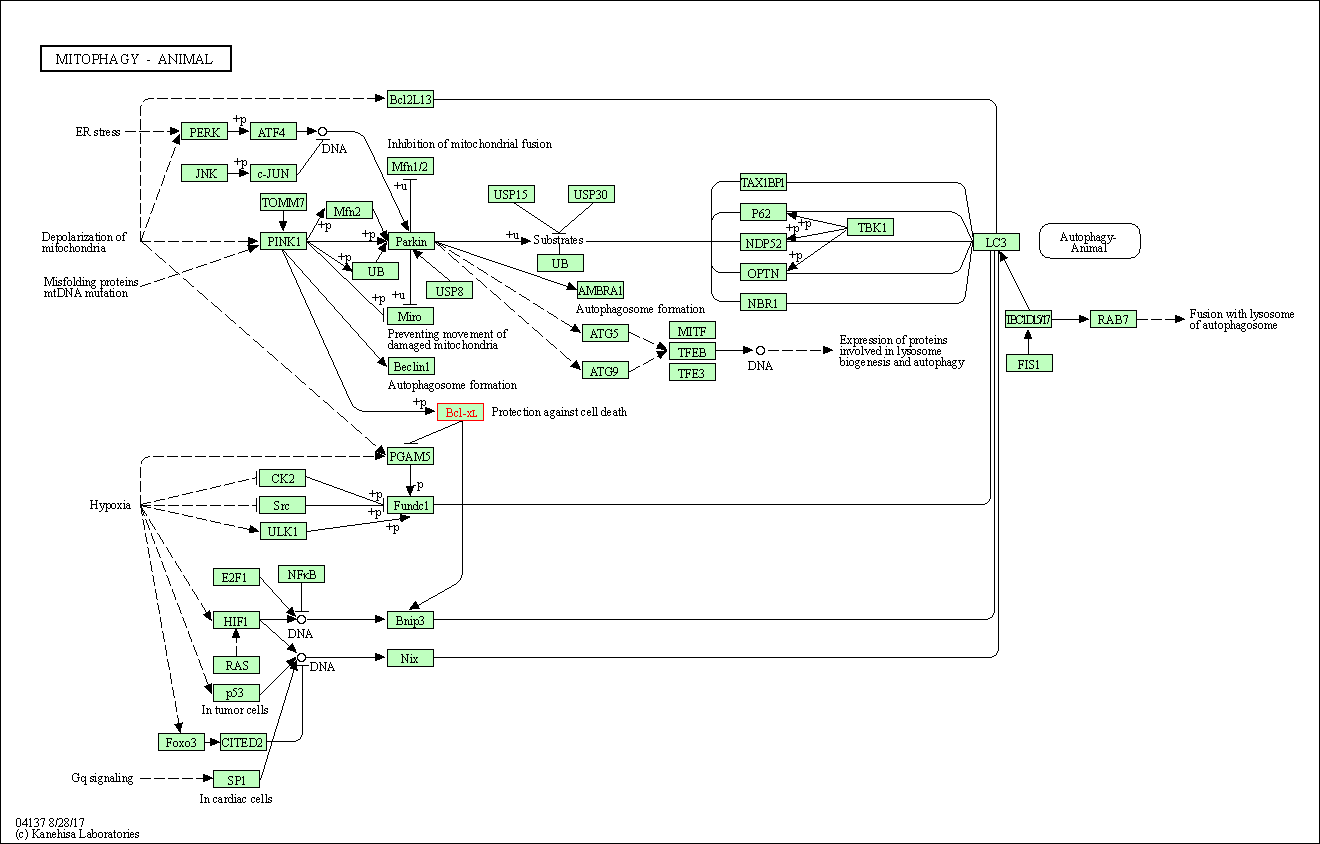
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Apoptosis - multiple species | hsa04215 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| JAK-STAT signaling pathway | hsa04630 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 50 | Degree centrality | 5.37E-03 | Betweenness centrality | 7.85E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.63E-01 | Radiality | 1.45E+01 | Clustering coefficient | 8.16E-02 |
| Neighborhood connectivity | 3.36E+01 | Topological coefficient | 3.55E-02 | Eccentricity | 10 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Phase II study of obatoclax mesylate (GX15-070), a small-molecule BCL-2 family antagonist, for patients with myelofibrosis. Clin Lymphoma Myeloma Leuk. 2010 Aug;10(4):285-9. | |||||
| REF 2 | ClinicalTrials.gov (NCT04472598) Study of Oral Navitoclax Tablet In Combination With Oral Ruxolitinib Tablet When Compared With Oral Ruxolitinib Tablet To Assess Change In Spleen Volume In Adult Participants With Myelofibrosis (TRANSFORM-1). U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT04865419) Study of AZD0466 Monotherapy or in Combination in Patients With Advanced Haematological Malignancies. U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT04210037) Study of APG-1252 Plus Paclitaxel in Patients With Relapsed/Refractory Small Cell Lung Cancer. U.S. National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of Roche (2009). | |||||
| REF 6 | ABT-263 and rapamycin act cooperatively to kill lymphoma cells in vitro and in vivo. Mol Cancer Ther. 2008 Oct;7(10):3265-74. | |||||
| REF 7 | ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008 May 1;68(9):3421-8. | |||||
| REF 8 | Mcl-1 inhibitors: a patent review.Expert Opin Ther Pat. 2017 Feb;27(2):163-178. | |||||
| REF 9 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2845). | |||||
| REF 11 | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations. Nat Commun. 2019 Jun 3;10(1):2385. | |||||
| REF 12 | An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005 Jun 2;435(7042):677-81. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

