Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T59845
(Former ID: TTDR00717)
|
|||||
| Target Name |
Serine/threonine PP1-alpha (PPP1CA)
|
|||||
| Synonyms |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit; Protein phosphatase 1alpha; PPP1A; PP-1A
Click to Show/Hide
|
|||||
| Gene Name |
PPP1CA
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Molluscum contagiosum [ICD-11: 1E76] | |||||
| Function |
Protein phosphatase 1 (PP1) is essential for cell division, and participates in the regulation of glycogen metabolism, muscle contractility and protein synthesis. Involved in regulation of ionic conductances and long-term synaptic plasticity. May play an important role in dephosphorylating substrates such as the postsynaptic density-associated Ca(2+)/calmodulin dependent protein kinase II. Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell cycle progression during the transition from mitosis into interphase. Regulates NEK2 function in terms of kinase activity and centrosome number and splitting, both in the presence and absence of radiation-induced DNA damage. Regulator of neural tube and optic fissure closure, and enteric neural crest cell (ENCCs) migration during development. In balance with CSNK1D and CSNK1E, determines the circadian period length, through the regulation of the speed and rhythmicity of PER1 and PER2 phosphorylation. May dephosphorylate CSNK1D and CSNK1E. Dephosphorylates the 'Ser-418' residue of FOXP3 in regulatory T-cells (Treg) from patients with rheumatoid arthritis, thereby inactivating FOXP3 and rendering Treg cells functionally defective. Dephosphorylates CENPA. Dephosphorylates the 'Ser-139' residue of ATG16L1 causing dissociation of ATG12-ATG5-ATG16L1 complex, thereby inhibiting autophagy. Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets.
Click to Show/Hide
|
|||||
| BioChemical Class |
Phosphoric monoester hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.1.3.16
|
|||||
| Sequence |
MSDSEKLNLDSIIGRLLEVQGSRPGKNVQLTENEIRGLCLKSREIFLSQPILLELEAPLK
ICGDIHGQYYDLLRLFEYGGFPPESNYLFLGDYVDRGKQSLETICLLLAYKIKYPENFFL LRGNHECASINRIYGFYDECKRRYNIKLWKTFTDCFNCLPIAAIVDEKIFCCHGGLSPDL QSMEQIRRIMRPTDVPDQGLLCDLLWSDPDKDVQGWGENDRGVSFTFGAEVVAKFLHKHD LDLICRAHQVVEDGYEFFAKRQLVTLFSAPNYCGEFDNAGAMMSVDETLMCSFQILKPAD KNKGKYGQFSGLNPGGRPITPPRNSAKAKK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Tautomycin diacid | Ligand Info | |||||
| Structure Description | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin inhibitor Tautomycin | PDB:3E7B | ||||
| Method | X-ray diffraction | Resolution | 1.70 Å | Mutation | No | [3] |
| PDB Sequence |
SLNLDSIIGR
15 LLEVQGSRPG25 KNVQLTENEI35 RGLCLKSREI45 FLSQPILLEL55 EAPLKICGDI 65 HGQYYDLLRL75 FEYGGFPPES85 NYLFLGDYVD95 RGKQSLETIC105 LLLAYKIKYP 115 ENFFLLRGNH125 ECASINRIYG135 FYDECKRRYN145 IKLWKTFTDC155 FNCLPIAAIV 165 DEKIFCCHGG175 LSPDLQSMEQ185 IRRIMRPTDV195 PDQGLLCDLL205 WSDPDKDVQG 215 WGENDRGVSF225 TFGAEVVAKF235 LHKHDLDLIC245 RAHQVVEDGY255 EFFAKRQLVT 265 LFSAPNYCGE275 FDNAGAMMSV285 DETLMCSFQI295 LKPA
|
|||||
|
|
ARG96
2.855
ASN124
4.294
HIS125
4.406
CYS127
3.685
SER129
3.736
ILE130
3.537
ILE133
3.922
TYR134
3.494
ASP194
4.266
VAL195
3.023
PRO196
3.500
ASP197
3.381
CYS202
4.242
|
|||||
| Ligand Name: Tautomycetin diacid form | Ligand Info | |||||
| Structure Description | Crystal structure of Protein Phosphatase 1 bound to the natural inhibitor Tautomycetin | PDB:6ALZ | ||||
| Method | X-ray diffraction | Resolution | 2.21 Å | Mutation | No | [4] |
| PDB Sequence |
LNLDSIIGRL
16 LEVQGSRPGK26 NVQLTENEIR36 GLCLKSREIF46 LSQPILLELE56 APLKICGDIH 66 GQYYDLLRLF76 EYGGFPPESN86 YLFLGDYVDR96 GKQSLETICL106 LLAYKIKYPE 116 NFFLLRGNHE126 CASINRIYGF136 YDECKRRYNI146 KLWKTFTDCF156 NCLPIAAIVD 166 EKIFCCHGGL176 SPDLQSMEQI186 RRIMRPTDVP196 DQGLLCDLLW206 SDPDKDVQGW 216 GENDRGVSFT226 FGAEVVAKFL236 HKHDLDLICR246 AHQVVEDGYE256 FFAKRQLVTL 266 FSAPNYCGEF276 DNAGAMMSVD286 ETLMCSFQIL296 KPA
|
|||||
|
|
ASP64
4.977
ARG96
2.922
ASN124
4.659
HIS125
4.769
CYS127
1.765
ALA128
4.995
SER129
3.469
ILE130
4.058
TYR134
3.458
VAL195
3.447
PRO196
4.306
ASP197
3.829
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| mRNA surveillance pathway | hsa03015 | Affiliated Target |
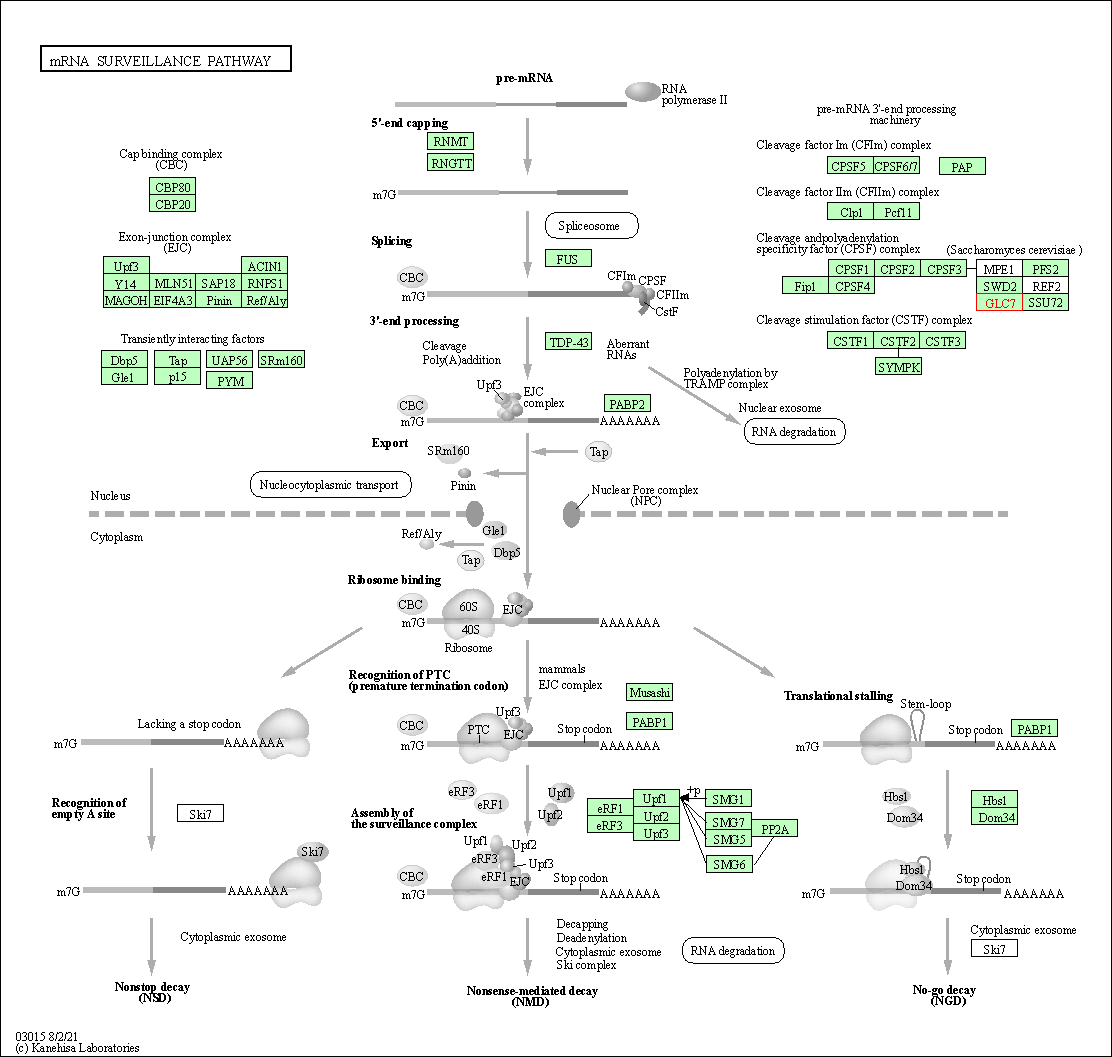
|
| Class: Genetic Information Processing => Translation | Pathway Hierarchy | ||
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |
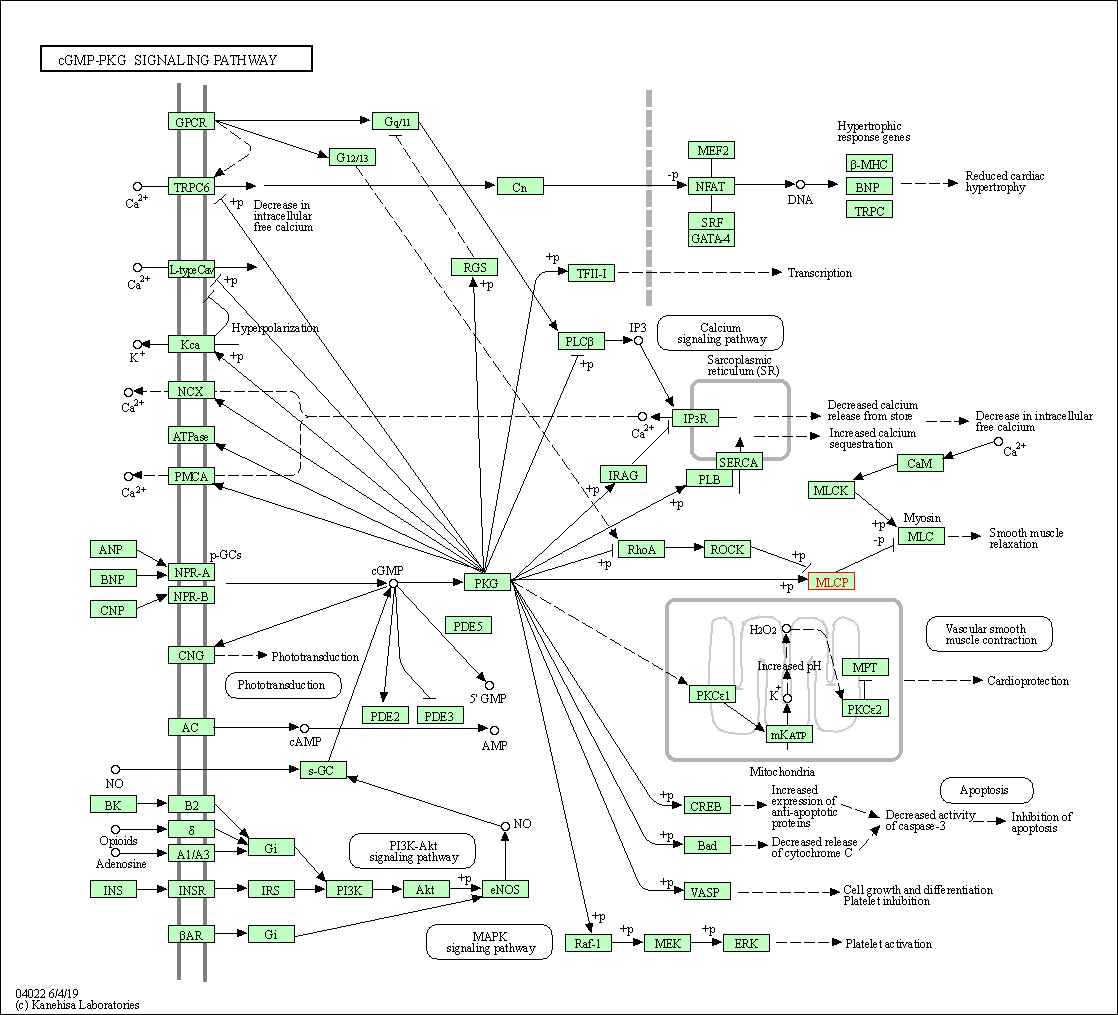
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |
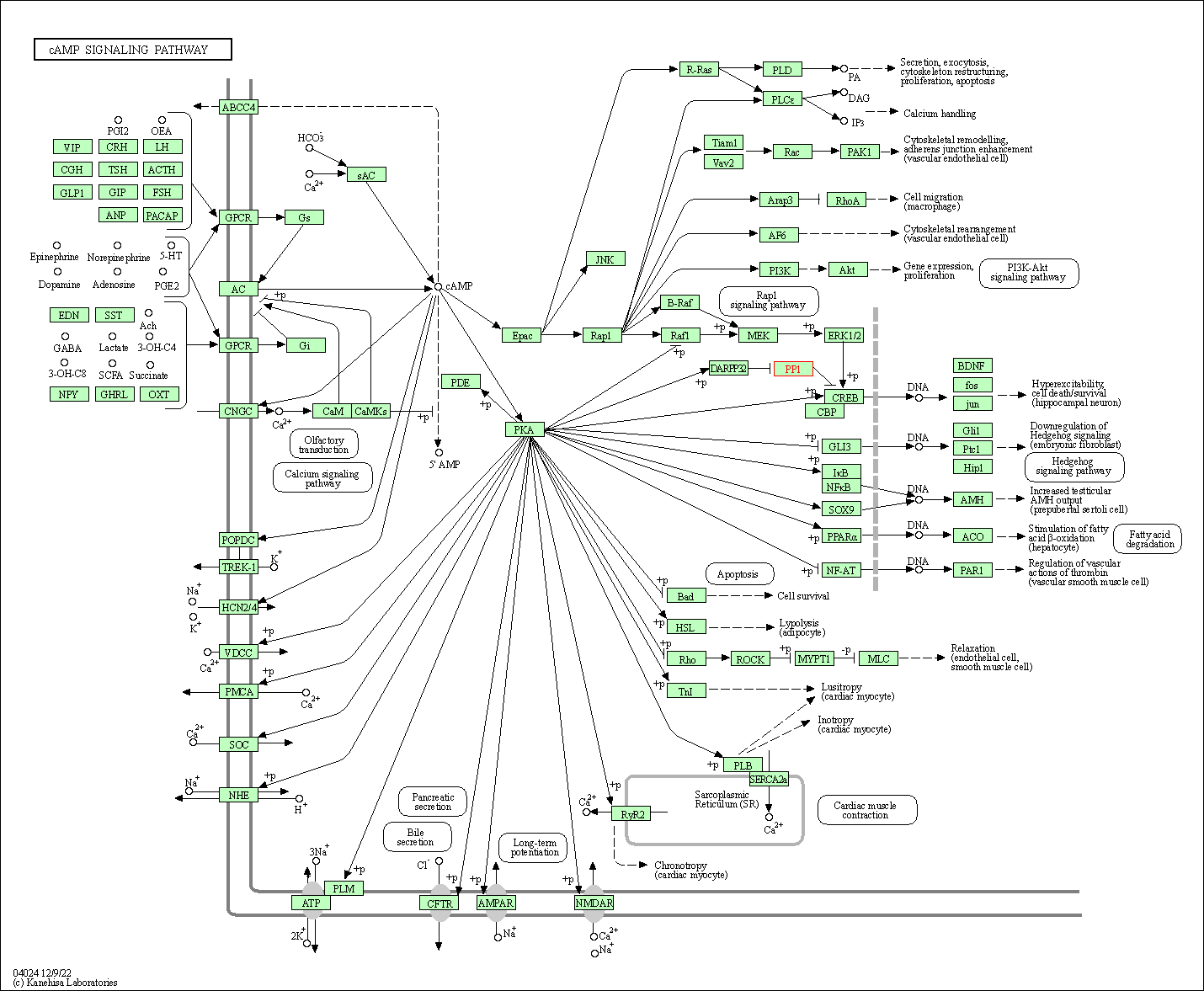
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |
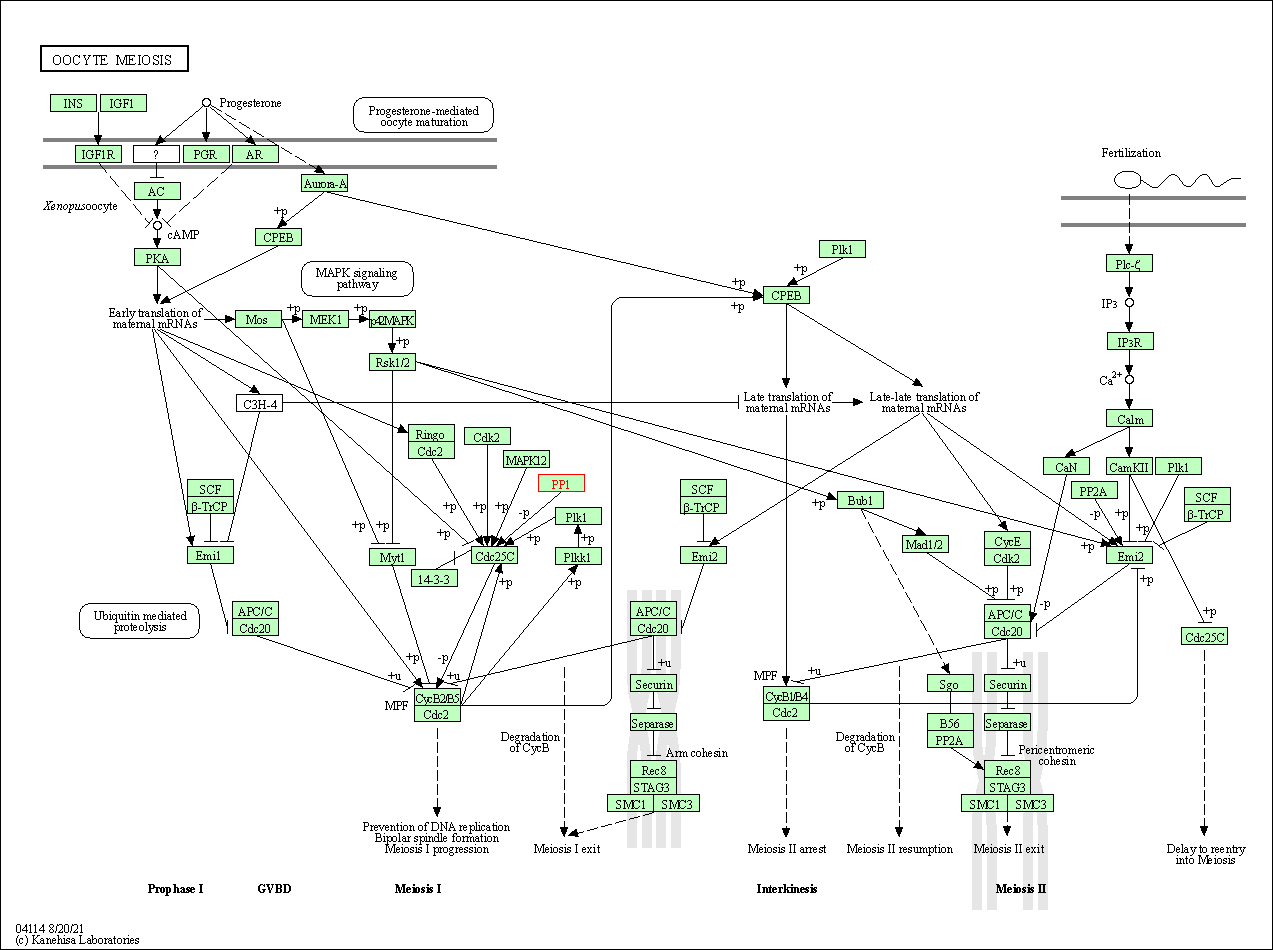
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Adrenergic signaling in cardiomyocytes | hsa04261 | Affiliated Target |
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Hippo signaling pathway | hsa04390 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 31 | Degree centrality | 3.33E-03 | Betweenness centrality | 1.69E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.24E-01 | Radiality | 1.39E+01 | Clustering coefficient | 1.12E-01 |
| Neighborhood connectivity | 1.57E+01 | Topological coefficient | 5.08E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Serine-threonine protein phosphatase inhibitors: development of potential therapeutic strategies. J Med Chem. 2002 Mar 14;45(6):1151-75. | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 3 | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol. 2009 Jan 9;385(1):11-21. | |||||
| REF 4 | PP1:Tautomycetin Complex Reveals a Path toward the Development of PP1-Specific Inhibitors. J Am Chem Soc. 2017 Dec 13;139(49):17703-17706. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

