Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T70176
(Former ID: TTDC00088)
|
|||||
| Target Name |
Cyclin-dependent kinase 2 (CDK2)
|
|||||
| Synonyms |
Sin3 associated polypeptide; SIN3-associated protein; P33 protein kinase; Cell division protein kinase 2; CDKN2
Click to Show/Hide
|
|||||
| Gene Name |
CDK2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 8 Target-related Diseases | + | ||||
| 1 | Thymoma [ICD-11: 2C27] | |||||
| 2 | Brain cancer [ICD-11: 2A00] | |||||
| 3 | Breast cancer [ICD-11: 2C60-2C6Y] | |||||
| 4 | Lung cancer [ICD-11: 2C25] | |||||
| 5 | Lymphoma [ICD-11: 2A80-2A86] | |||||
| 6 | Malignant haematopoietic neoplasm [ICD-11: 2B33] | |||||
| 7 | Non-small-cell lung cancer [ICD-11: 2C25] | |||||
| 8 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1. Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability. Phosphorylates CDK2AP2. Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.22
|
|||||
| Sequence |
MENFQKVEKIGEGTYGVVYKARNKLTGEVVALKKIRLDTETEGVPSTAIREISLLKELNH
PNIVKLLDVIHTENKLYLVFEFLHQDLKKFMDASALTGIPLPLIKSYLFQLLQGLAFCHS HRVLHRDLKPQNLLINTEGAIKLADFGLARAFGVPVRTYTHEVVTLWYRAPEILLGCKYY STAVDIWSLGCIFAEMVTRRALFPGDSEIDQLFRIFRTLGTPDEVVWPGVTSMPDYKPSF PKWARQDFSKVVPPLDEDGRSLLSQMLHYDPNKRISAKAALAHPFFQDVTKPVPHLRL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T75H3W | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 11 Clinical Trial Drugs | + | ||||
| 1 | PHA848125 | Drug Info | Phase 2 | Thymic cancer | [2], [3] | |
| 2 | R-roscovitine | Drug Info | Phase 2 | Non-small-cell lung cancer | [4], [5] | |
| 3 | Ro 31-7453 | Drug Info | Phase 2 | Solid tumour/cancer | [6] | |
| 4 | TG02 | Drug Info | Phase 1/2 | Anaplastic astrocytoma | [8], [9] | |
| 5 | AG-024322 | Drug Info | Phase 1 | Solid tumour/cancer | [13] | |
| 6 | AT7519 | Drug Info | Phase 1 | Solid tumour/cancer | [14] | |
| 7 | AZD-5438 | Drug Info | Phase 1 | Solid tumour/cancer | [15], [16] | |
| 8 | CYC065 | Drug Info | Phase 1 | Lymphoma | [8] | |
| 9 | PHA-793887 | Drug Info | Phase 1 | Solid tumour/cancer | [17] | |
| 10 | RGB-286638 | Drug Info | Phase 1 | Haematological malignancy | [18], [19] | |
| 11 | SNS-032 | Drug Info | Phase 1 | Solid tumour/cancer | [20], [21] | |
| Discontinued Drug(s) | [+] 4 Discontinued Drugs | + | ||||
| 1 | SCH 727965 | Drug Info | Discontinued in Phase 3 | Acute lymphoblastic leukaemia | [22], [23], [24] | |
| 2 | BAY 10-00394 | Drug Info | Discontinued in Phase 2 | Small-cell lung cancer | [25], [26] | |
| 3 | R547 | Drug Info | Discontinued in Phase 1 | Advanced solid tumour | [27], [28] | |
| 4 | ZK 304709 | Drug Info | Discontinued in Phase 1 | Advanced solid tumour | [29] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | INOC-005 | Drug Info | Preclinical | Solid tumour/cancer | [31] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 117 Inhibitor drugs | + | ||||
| 1 | PHA848125 | Drug Info | [33], [34] | |||
| 2 | R-roscovitine | Drug Info | [13], [35], [36] | |||
| 3 | Ro 31-7453 | Drug Info | [35] | |||
| 4 | AG-024322 | Drug Info | [13] | |||
| 5 | AT7519 | Drug Info | [38] | |||
| 6 | CYC065 | Drug Info | [8] | |||
| 7 | PHA-793887 | Drug Info | [17] | |||
| 8 | RGB-286638 | Drug Info | [40] | |||
| 9 | SNS-032 | Drug Info | [21], [41] | |||
| 10 | 4-(thiazol-5-yl)-pyrimidine derivative 1 | Drug Info | [42] | |||
| 11 | 4-(thiazol-5-yl)-pyrimidine derivative 2 | Drug Info | [42] | |||
| 12 | 4-amino-3,5-di-substituted-thiazole derivative 1 | Drug Info | [42] | |||
| 13 | Alkyl sulfone derivative 1 | Drug Info | [42] | |||
| 14 | Aniline derivative 1 | Drug Info | [42] | |||
| 15 | Diamidothiazole derivative 1 | Drug Info | [42] | |||
| 16 | Diaryl amine derivative 1 | Drug Info | [42] | |||
| 17 | Flavopiridol analog 1 | Drug Info | [42] | |||
| 18 | Fluorinated compound 1 | Drug Info | [42] | |||
| 19 | Indole-based analog 11 | Drug Info | [42] | |||
| 20 | Indole-based analog 13 | Drug Info | [42] | |||
| 21 | Isosteric imidazolyl pyrimidine derivative 1 | Drug Info | [42] | |||
| 22 | N-(pyridin-2-yl)pyridine methylsulfone derivative 1 | Drug Info | [42] | |||
| 23 | N-(pyridin-2-yl)pyrimidin-4-amine derivative 2 | Drug Info | [42] | |||
| 24 | N-phenyl-pyrimidin-4-amine derivative 1 | Drug Info | [42] | |||
| 25 | Naphthyridine and isoquinoline derivative 1 | Drug Info | [42] | |||
| 26 | Nitrogen mustard derivative 1 | Drug Info | [42] | |||
| 27 | Oxazolyl methylthiothiazole derivative 1 | Drug Info | [42] | |||
| 28 | Palbociclib/ribociclib analog 1 | Drug Info | [42] | |||
| 29 | PMID25991433-Compound-A1 | Drug Info | [43] | |||
| 30 | PMID26161698-Compound-25 | Drug Info | [42] | |||
| 31 | Pyrazolo-triazine derivative 2 | Drug Info | [42] | |||
| 32 | Pyrazolo[1,5-a]-1,3,5-triazine derivative 1 | Drug Info | [42] | |||
| 33 | Pyrrolo[2,3-d]pyrimidine derivative 9 | Drug Info | [42] | |||
| 34 | Roscovitine derivative 1 | Drug Info | [42] | |||
| 35 | SCH 727965 | Drug Info | [13] | |||
| 36 | BAY 10-00394 | Drug Info | [44] | |||
| 37 | R547 | Drug Info | [13] | |||
| 38 | ZK 304709 | Drug Info | [13] | |||
| 39 | INOC-005 | Drug Info | [45] | |||
| 40 | Olomoucine | Drug Info | [46] | |||
| 41 | (2'Z,3'E)-5-Chloro-5'-chloro-indirubin-3'-oxime | Drug Info | [47] | |||
| 42 | (2'Z,3'E)-5-Chloro-5'-fluoro-indirubin-3'-oxime | Drug Info | [47] | |||
| 43 | (2'Z,3'E)-5-Chloro-5'-hydroxy-indirubin-3'-oxime | Drug Info | [47] | |||
| 44 | (2'Z,3'E)-5-Chloro-5'-methyl-indirubin-3'-oxime | Drug Info | [47] | |||
| 45 | (2'Z,3'E)-5-Fluoro-5'-chloro-indirubin-3'-oxime | Drug Info | [47] | |||
| 46 | (2'Z,3'E)-5-Fluoro-5'-fluoro-indirubin-3'-oxime | Drug Info | [47] | |||
| 47 | (2'Z,3'E)-5-Fluoro-5'-hydroxy-indirubin-3'-oxime | Drug Info | [47] | |||
| 48 | (2'Z,3'E)-5-Fluoro-5'-methoxy-indirubin-3'-oxime | Drug Info | [47] | |||
| 49 | (2'Z,3'E)-5-Nitro-5'-chloro-indirubin-3'-oxime | Drug Info | [47] | |||
| 50 | (2'Z,3'E)-5-Nitro-5'-fluoro-indirubin-3'-oxime | Drug Info | [47] | |||
| 51 | (2'Z,3'E)-5-Nitro-5'-methoxy-indirubin-3'-oxime | Drug Info | [47] | |||
| 52 | (2'Z,3'E)-5-Nitro-5'-methyl-indirubin-3'-oxime | Drug Info | [47] | |||
| 53 | 1-Amino-6-Cyclohex-3-Enylmethyloxypurine | Drug Info | [48] | |||
| 54 | 10Z-Hymenialdisine | Drug Info | [46] | |||
| 55 | 2-((3,5-diamino-1H-pyrazol-4-yl)diazenyl)phenol | Drug Info | [49] | |||
| 56 | 2-ANILINO-6-CYCLOHEXYLMETHOXYPURINE | Drug Info | [50], [51] | |||
| 57 | 3,4-di-(4-methoxyphenyl)-1H-pyrrole-2,5-dione | Drug Info | [52] | |||
| 58 | 3,4-diphenyl-1H-pyrrole-2,5-dione | Drug Info | [52] | |||
| 59 | 3-(4-methoxyphenyl)-4-phenyl-1H-pyrrole-2,5-dione | Drug Info | [52] | |||
| 60 | 3-(indole-3-yl)-4-phenyl-1H-pyrrole-2,5-dione | Drug Info | [52] | |||
| 61 | 4-(2,4-Dimethyl-Thiazol-5-Yl)-Pyrimidin-2-Ylamine | Drug Info | [48] | |||
| 62 | 4-[(3,5-diamino-1H-pyrazol-4-yl)diazenyl]phenol | Drug Info | [49] | |||
| 63 | 4-[(6-chloropyrazin-2-yl)amino]benzenesulfonamide | Drug Info | [50] | |||
| 64 | 4-[3-Hydroxyanilino]-6,7-Dimethoxyquinazoline | Drug Info | [53] | |||
| 65 | 5-hydroxynaphthalene-1-sulfonamide | Drug Info | [50] | |||
| 66 | 5-nitroindirubin-3'-oxime | Drug Info | [47] | |||
| 67 | 6-(3-Amino-benzyloxy)-9H-purin-2-ylamine | Drug Info | [54] | |||
| 68 | 6-(3-Methyl-benzyloxy)-9H-purin-2-ylamine | Drug Info | [54] | |||
| 69 | 6-(Cyclohex-3-enylmethoxy)-9H-purin-2-ylamine | Drug Info | [54] | |||
| 70 | 6-CYCLOHEXYLMETHOXY-2-(3'-CHLOROANILINO) PURINE | Drug Info | [50] | |||
| 71 | 6-Cyclohexylmethoxy-pyrimidine-2,4,5-triamine | Drug Info | [55] | |||
| 72 | 6-O-Cyclohexylmethyl Guanine | Drug Info | [48], [56] | |||
| 73 | 9-Nitropaullone | Drug Info | [46] | |||
| 74 | aloisine | Drug Info | [57] | |||
| 75 | aloisine A | Drug Info | [58] | |||
| 76 | aminopurvalanol A | Drug Info | [59] | |||
| 77 | Benzyl-(9-isopropyl-9H-purin-6-yl)-amine | Drug Info | [60] | |||
| 78 | BMS-265246 | Drug Info | [61] | |||
| 79 | BMS-536924 | Drug Info | [62] | |||
| 80 | BOHEMINE | Drug Info | [60] | |||
| 81 | BX-795 | Drug Info | [63] | |||
| 82 | BX-912 | Drug Info | [63] | |||
| 83 | Cdk1/2 inhibitor III | Drug Info | [64] | |||
| 84 | Cdk4 inhibitor III | Drug Info | [65] | |||
| 85 | CVT-313 | Drug Info | [66] | |||
| 86 | Double Oxidized Cysteine | Drug Info | [48] | |||
| 87 | GW-8510 | Drug Info | [67] | |||
| 88 | Indirubin-5-sulfonate | Drug Info | [46] | |||
| 89 | JNJ-7706621 | Drug Info | [68] | |||
| 90 | K00024 | Drug Info | [69] | |||
| 91 | Lysine Nz-Carboxylic Acid | Drug Info | [48] | |||
| 92 | MERIOLIN 1 | Drug Info | [70] | |||
| 93 | MERIOLIN 2 | Drug Info | [70] | |||
| 94 | MERIOLIN 3 | Drug Info | [70] | |||
| 95 | MERIOLIN 4 | Drug Info | [70] | |||
| 96 | MERIOLIN 5 | Drug Info | [70] | |||
| 97 | MERIOLIN 6 | Drug Info | [70] | |||
| 98 | MERIOLIN 8 | Drug Info | [70] | |||
| 99 | N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE | Drug Info | [50] | |||
| 100 | N-(4-amino-5-cyano-6-phenylpyridin-2-yl)acetamide | Drug Info | [71] | |||
| 101 | N-(4-sulfamoylphenyl)-1H-indazole-3-carboxamide | Drug Info | [50] | |||
| 102 | N-(5-Cyclopropyl-1h-Pyrazol-3-Yl)Benzamide | Drug Info | [48] | |||
| 103 | N-phenyl-1H-pyrazole-3-carboxamide | Drug Info | [50] | |||
| 104 | NU6140 | Drug Info | [72] | |||
| 105 | Oxindole 95 | Drug Info | [46] | |||
| 106 | PF-228 | Drug Info | [73] | |||
| 107 | PHA-690509 | Drug Info | [74] | |||
| 108 | PHENYLAMINOIMIDAZO(1,2-ALPHA)PYRIDINE | Drug Info | [50] | |||
| 109 | PMID18986805C9b | Drug Info | [75] | |||
| 110 | PMID19115845C89S | Drug Info | [76] | |||
| 111 | Purvalanol A | Drug Info | [77] | |||
| 112 | PYRAZOLOPYRIDAZINE 1 | Drug Info | [78] | |||
| 113 | PYRAZOLOPYRIDAZINE 2 | Drug Info | [78] | |||
| 114 | RESCOVITINE | Drug Info | [79] | |||
| 115 | SCH-546909 | Drug Info | [66] | |||
| 116 | SU9516 | Drug Info | [46] | |||
| 117 | TRIAZOLOPYRIMIDINE | Drug Info | [50] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | TG02 | Drug Info | [37] | |||
| 2 | AZD-5438 | Drug Info | [39], [16] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Sunitinib | Ligand Info | |||||
| Structure Description | CDK2 in complex with SUNITINIB | PDB:3TI1 | ||||
| Method | X-ray diffraction | Resolution | 1.99 Å | Mutation | No | [80] |
| PDB Sequence |
MENFQKVEKI
10 GEGTYGVVYK20 ARNKLTGEVV30 ALKKIRTEGV44 PSTAIREISL54 LKELNHPNIV 64 KLLDVIHTEN74 KLYLVFEFLH84 QDLKKFMDAS94 ALTGIPLPLI104 KSYLFQLLQG 114 LAFCHSHRVL124 HRDLKPQNLL134 INTEGAIKLA144 DFGLARAFGV154 PVRTYTHEVV 164 TLWYRAPEIL174 LGCKYYSTAV184 DIWSLGCIFA194 EMVTRRALFP204 GDSEIDQLFR 214 IFRTLGTPDE224 VVWPGVTSMP234 DYKPSFPKWA244 RQDFSKVVPP254 LDEDGRSLLS 264 QMLHYDPNKR274 ISAKAALAHP284 FFQDVTKPVP294 HLRL
|
|||||
|
|
||||||
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with ATP | PDB:4EOJ | ||||
| Method | X-ray diffraction | Resolution | 1.65 Å | Mutation | Yes | [81] |
| PDB Sequence |
GSMENFQKVE
8 KIGEGTYGVV18 YKARNKLTGE28 VVALKKIRLE42 GVPSTAIREI52 SLLKELNHPN 62 IVKLLDVIHT72 ENKLYLVFEF82 LSMDLKDFMD92 ASALTGIPLP102 LIKSYLFQLL 112 QGLAFCHSHR122 VLHRDLKPQN132 LLINTEGAIK142 LADFGLARAF152 GVPVRTYHEV 163 VTLWYRAPEI173 LLGCKYYSTA183 VDIWSLGCIF193 AEMVTRRALF203 PGDSEIDQLF 213 RIFRTLGTPD223 EVVWPGVTSM233 PDYKPSFPKW243 ARQDFSKVVP253 PLDEDGRSLL 263 SQMLHYDPNK273 RISAKAALAH283 PFFQDVTKPV293 PHLRL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| FoxO signaling pathway | hsa04068 | Affiliated Target |
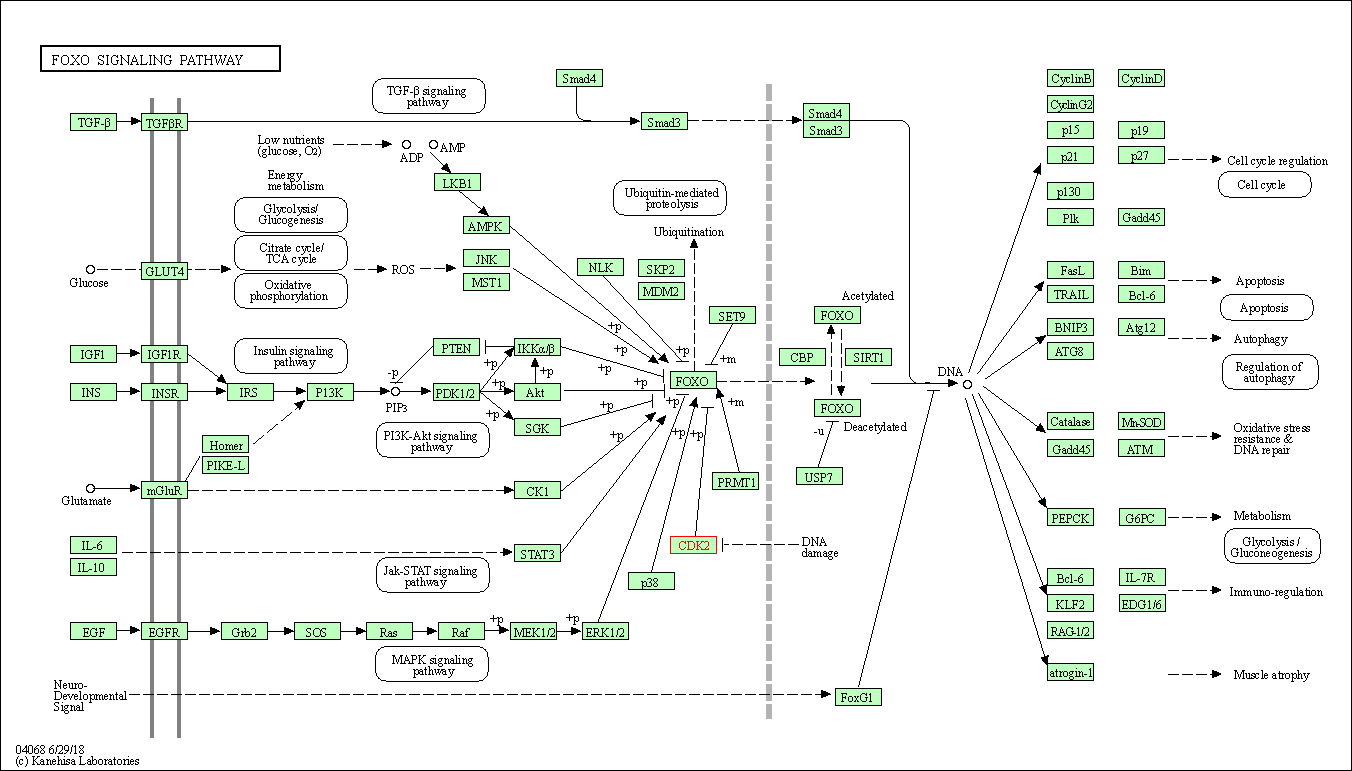
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cell cycle | hsa04110 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| p53 signaling pathway | hsa04115 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
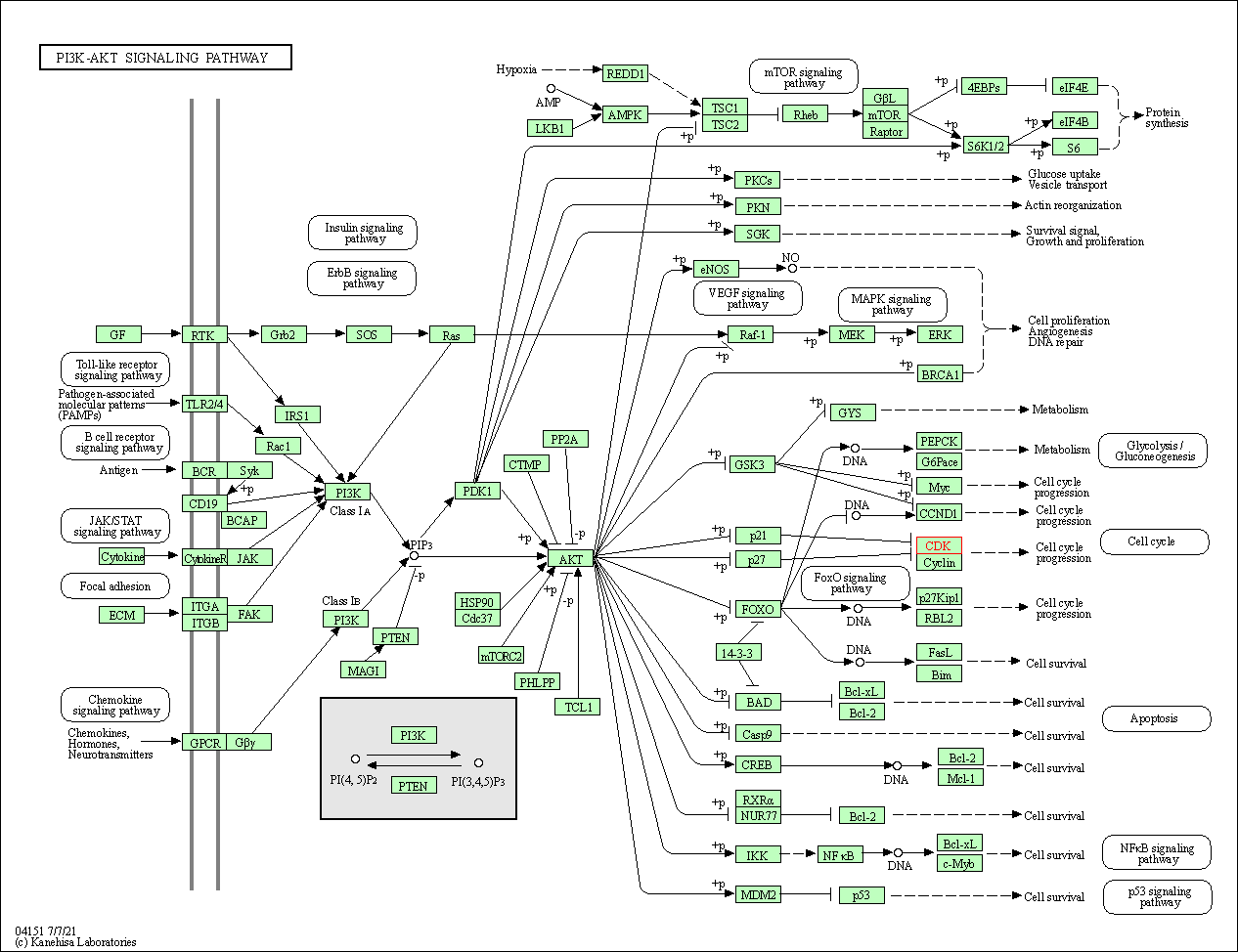
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |
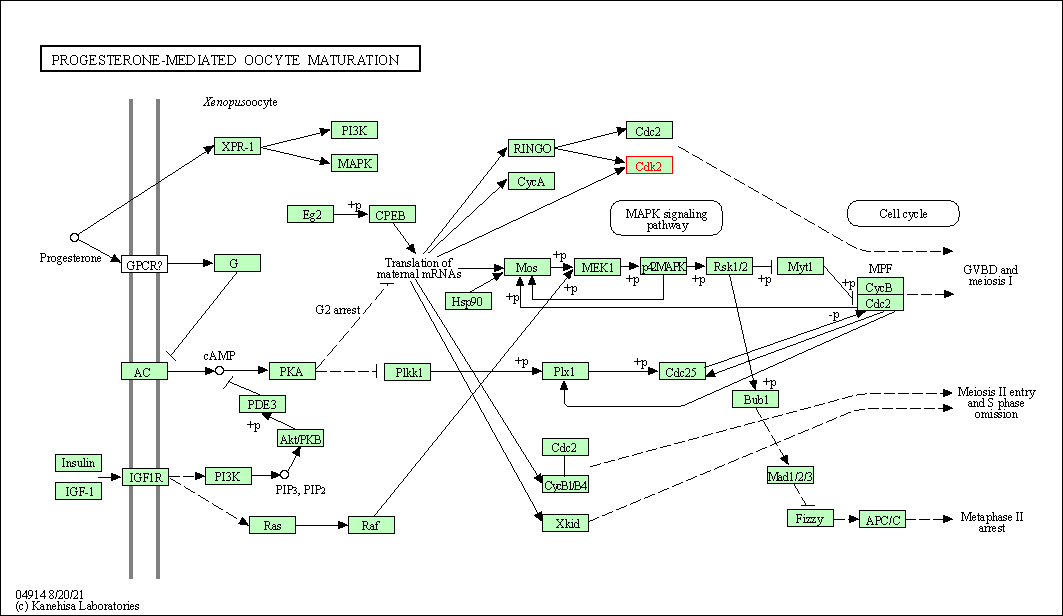
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 74 | Degree centrality | 7.95E-03 | Betweenness centrality | 4.53E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.60E-01 | Radiality | 1.45E+01 | Clustering coefficient | 1.78E-01 |
| Neighborhood connectivity | 4.41E+01 | Topological coefficient | 4.18E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Agreement signed with Prostagenics to develop prostate cancer treatment. Innovate Oncology, Inc. 2005. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7938). | |||||
| REF 3 | ClinicalTrials.gov (NCT01011439) Phase II Study Of Oral PHA-848125AC In Patients With Thymic Carcinoma. U.S. National Institutes of Health. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6035). | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010117) | |||||
| REF 6 | A phase 2 study of oral MKC-1, an inhibitor of importin-beta, tubulin, and the mTOR pathway in patients with unresectable or metastatic pancreatic cancer. Invest New Drugs. 2012 Aug;30(4):1614-20. | |||||
| REF 7 | ClinicalTrials.gov (NCT04541225) Phase 1/2 Dose Escalation, Safety, Pharmacokinetics, and Efficacy Study of NUV-422 in Adults With Recurrent or Refractory High-grade Gliomas and Solid Tumors. U.S.National Institutes of Health. | |||||
| REF 8 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 9 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 10 | ClinicalTrials.gov (NCT04553133) PF-07104091 as a Single Agent and in Combination Therapy. U.S. National Institutes of Health. | |||||
| REF 11 | ClinicalTrials.gov (NCT03690154) A Phase 1 Study to Evaluate FN-1501 Monotherapy in Patients With Advanced Solid Tumors and R/R AML. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT05304962) First-in-Human, Escalating Oral Dose Study of RGT-419B Given Alone and With Endocrine Therapy in Subjects With Hormone Receptor Positive, Human Epidermal Growth Factor Receptor 2 Negative Advanced/Metastatic Breast Cancer. U.S.National Institutes of Health. | |||||
| REF 13 | Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009 Jul;8(7):547-66. | |||||
| REF 14 | ClinicalTrials.gov (NCT02503709) Onalespib and CDKI AT7519 in Treating Patients With Solid Tumors That Are Metastatic or Cannot Be Removed by Surgery in National Cancer Institute (NCI). | |||||
| REF 15 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8473). | |||||
| REF 16 | AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol Cancer Ther. 2009 Jul;8(7):1856-66. | |||||
| REF 17 | A first in man, phase I dose-escalation study of PHA-793887, an inhibitor of multiple cyclin-dependent kinases (CDK2, 1 and 4) reveals unexpected h... Cell Cycle. 2011 Mar 15;10(6):963-70. | |||||
| REF 18 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7744). | |||||
| REF 19 | ClinicalTrials.gov (NCT01168882) Safety and Tolerability of RGB-286638 in Patients With Selected, Relapsed or Refractory Hematological Malignancies. U.S. National Institutes of Health. | |||||
| REF 20 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5670). | |||||
| REF 21 | Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009 May 7;113(19):4637-45. | |||||
| REF 22 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7379). | |||||
| REF 23 | Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood. 2015 Jan 15;125(3):443-8. | |||||
| REF 24 | Cyclin-dependent kinase inhibitor Dinaciclib (SCH727965) inhibits pancreatic cancer growth and progression in murine xenograft models. Cancer Biol Ther. 2011 Oct 1;12(7):598-609. | |||||
| REF 25 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7874). | |||||
| REF 26 | BAY 1000394, a novel cyclin-dependent kinase inhibitor, with potent antitumor activity in mono- and in combination treatment upon oral application. Mol Cancer Ther. 2012 Oct;11(10):2265-73. | |||||
| REF 27 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5707). | |||||
| REF 28 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800018924) | |||||
| REF 29 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800022386) | |||||
| REF 30 | Emerging drugs for obesity: linking novel biological mechanisms to pharmaceutical pipelines. Expert Opin Emerg Drugs. 2005 Aug;10(3):643-60. | |||||
| REF 31 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800022337) | |||||
| REF 32 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800014130) | |||||
| REF 33 | Identification of N,1,4,4-tetramethyl-8-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxamide (PHA-848125), a potent, orally available cyclin dependent kinase inhibitor. J Med Chem. 2009 Aug 27;52(16):5152-63. | |||||
| REF 34 | The cyclin-dependent kinase inhibitor PHA-848125 suppresses the in vitro growth of human melanomas sensitive or resistant to temozolomide, and shows synergistic effects in combination with this triazene compound. Pharmacol Res. 2010 May;61(5):437-48. | |||||
| REF 35 | A comparison of physicochemical property profiles of marketed oral drugs and orally bioavailable anti-cancer protein kinase inhibitors in clinical development. Curr Top Med Chem. 2007;7(14):1408-22. | |||||
| REF 36 | Cdk2 plays a critical role in hepatocyte cell cycle progression and survival in the setting of cyclin D1 expression in vivo. Cell Cycle. 2009 Sep 1;8(17):2802-9. | |||||
| REF 37 | Preclinical metabolism and pharmacokinetics of SB1317 (TG02), a potent CDK/JAK2/FLT3 inhibitor. Drug Metab Lett. 2012 Mar;6(1):33-42. | |||||
| REF 38 | Biological characterization of AT7519, a small-molecule inhibitor of cyclin-dependent kinases, in human tumor cell lines. Mol Cancer Ther. 2009 Feb;8(2):324-32. | |||||
| REF 39 | AZD5438, an inhibitor of Cdk1, 2, and 9, enhances the radiosensitivity of non-small cell lung carcinoma cells.Int J Radiat Oncol Biol Phys.2012 Nov 15;84(4):e507-14. | |||||
| REF 40 | Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia. 2013 Dec;27(12):2366-75. | |||||
| REF 41 | Development of cell-cycle inhibitors for cancer therapy. Curr Oncol. 2009 Mar;16(2):36-43. | |||||
| REF 42 | Cyclin-dependent kinase inhibitors for cancer therapy: a patent review (2009 - 2014).Expert Opin Ther Pat. 2015;25(9):953-70. | |||||
| REF 43 | c-Jun N-terminal kinase inhibitors: a patent review (2010 - 2014).Expert Opin Ther Pat. 2015;25(8):849-72. | |||||
| REF 44 | National Cancer Institute Drug Dictionary (drug id 770319). | |||||
| REF 45 | What are next generation innovative therapeutic targets. J Pharmacol Exp Ther. 2009 Jul;330(1):304-15. | |||||
| REF 46 | Pharmacological inhibitors of cyclin-dependent kinases. Trends Pharmacol Sci. 2002 Sep;23(9):417-25. | |||||
| REF 47 | 5,5'-substituted indirubin-3'-oxime derivatives as potent cyclin-dependent kinase inhibitors with anticancer activity. J Med Chem. 2010 May 13;53(9):3696-706. | |||||
| REF 48 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 49 | 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J Med Chem. 2006 Nov 2;49(22):6500-9. | |||||
| REF 50 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 51 | Halogen bonding--a novel interaction for rational drug design J Med Chem. 2009 May 14;52(9):2854-62. | |||||
| REF 52 | Design, synthesis, and biological evaluation of 3,4-diarylmaleimides as angiogenesis inhibitors. J Med Chem. 2006 Feb 23;49(4):1271-81. | |||||
| REF 53 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 54 | Probing the ATP ribose-binding domain of cyclin-dependent kinases 1 and 2 with O(6)-substituted guanine derivatives. J Med Chem. 2002 Aug 1;45(16):3381-93. | |||||
| REF 55 | 4-Alkoxy-2,6-diaminopyrimidine derivatives: inhibitors of cyclin dependent kinases 1 and 2. Bioorg Med Chem Lett. 2003 Jan 20;13(2):217-22. | |||||
| REF 56 | Novel structural features of CDK inhibition revealed by an ab initio computational method combined with dynamic simulations. J Med Chem. 2006 Aug 24;49(17):5141-53. | |||||
| REF 57 | Identification of binding specificity-determining features in protein families. J Med Chem. 2012 Mar 8;55(5):1926-39. | |||||
| REF 58 | Aloisines, a new family of CDK/GSK-3 inhibitors. SAR study, crystal structure in complex with CDK2, enzyme selectivity, and cellular effects. J Med Chem. 2003 Jan 16;46(2):222-36. | |||||
| REF 59 | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20523-8. | |||||
| REF 60 | Docking-based development of purine-like inhibitors of cyclin-dependent kinase-2. J Med Chem. 2000 Jun 29;43(13):2506-13. | |||||
| REF 61 | A robust high-content imaging approach for probing the mechanism of action and phenotypic outcomes of cell-cycle modulators. Mol Cancer Ther. 2011 Feb;10(2):242-54. | |||||
| REF 62 | Discovery of a (1H-benzoimidazol-2-yl)-1H-pyridin-2-one (BMS-536924) inhibitor of insulin-like growth factor I receptor kinase with in vivo antitum... J Med Chem. 2005 Sep 8;48(18):5639-43. | |||||
| REF 63 | Novel small molecule inhibitors of 3-phosphoinositide-dependent kinase-1. J Biol Chem. 2005 May 20;280(20):19867-74. | |||||
| REF 64 | 1-Acyl-1H-[1,2,4]triazole-3,5-diamine analogues as novel and potent anticancer cyclin-dependent kinase inhibitors: synthesis and evaluation of biological activities. J Med Chem. 2005 Jun 30;48(13):4208-11. | |||||
| REF 65 | 5-Arylamino-2-methyl-4,7-dioxobenzothiazoles as inhibitors of cyclin-dependent kinase 4 and cytotoxic agents. Bioorg Med Chem Lett. 2000 Mar 6;10(5):461-4. | |||||
| REF 66 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1973). | |||||
| REF 67 | Discovery and in vitro evaluation of potent TrkA kinase inhibitors: oxindole and aza-oxindoles. Bioorg Med Chem Lett. 2004 Feb 23;14(4):953-7. | |||||
| REF 68 | Synthesis and evaluation of N-acyl sulfonamides as potential prodrugs of cyclin-dependent kinase inhibitor JNJ-7706621. Bioorg Med Chem Lett. 2006 Jul 15;16(14):3639-41. | |||||
| REF 69 | Synthesis, structure-activity relationship, and biological studies of indolocarbazoles as potent cyclin D1-CDK4 inhibitors. J Med Chem. 2003 May 22;46(11):2027-30. | |||||
| REF 70 | Meriolins (3-(pyrimidin-4-yl)-7-azaindoles): synthesis, kinase inhibitory activity, cellular effects, and structure of a CDK2/cyclin A/meriolin com... J Med Chem. 2008 Feb 28;51(4):737-51. | |||||
| REF 71 | Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J Med Chem. 2006 Jun 15;49(12):3563-80. | |||||
| REF 72 | Potentiation of paclitaxel-induced apoptosis by the novel cyclin-dependent kinase inhibitor NU6140: a possible role for survivin down-regulation. Mol Cancer Ther. 2005 Sep;4(9):1328-37. | |||||
| REF 73 | Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007 May 18;282(20):14845-52. | |||||
| REF 74 | Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2010 Aug 17;107(33):14621-6. | |||||
| REF 75 | Imidazole pyrimidine amides as potent, orally bioavailable cyclin-dependent kinase inhibitors. Bioorg Med Chem Lett. 2008 Dec 15;18(24):6486-9. | |||||
| REF 76 | First Cdc7 kinase inhibitors: pyrrolopyridinones as potent and orally active antitumor agents. 2. Lead discovery. J Med Chem. 2009 Jan 22;52(2):293-307. | |||||
| REF 77 | The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007 Dec 15;408(3):297-315. | |||||
| REF 78 | N-Phenyl-4-pyrazolo[1,5-b]pyridazin-3-ylpyrimidin-2-amines as potent and selective inhibitors of glycogen synthase kinase 3 with good cellular effi... J Med Chem. 2004 Sep 9;47(19):4716-30. | |||||
| REF 79 | Design, synthesis, and biological evaluation of novel pyrimidine derivatives as CDK2 inhibitors. Eur J Med Chem. 2010 Mar;45(3):1158-66. | |||||
| REF 80 | A novel approach to the discovery of small-molecule ligands of CDK2. Chembiochem. 2012 Sep 24;13(14):2128-36. | |||||
| REF 81 | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity. Chem Biol. 2012 Aug 24;19(8):1028-40. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

