Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T78319
(Former ID: TTDR00525)
|
|||||
| Target Name |
Extracellular matrix receptor III (CD44)
|
|||||
| Synonyms |
Phagocytic glycoprotein I; Phagocytic glycoprotein 1; PGP-I; PGP-1; MIC4; MDU3; MDU2; LHR; Hyaluronate receptor; Hermes antigen; Heparan sulfate proteoglycan; HUTCH-I; GP90 lymphocyte homing/adhesion receptor; Extracellular matrix receptor-III; Epican; ECMR-III; CDw44; CD44 antigen
Click to Show/Hide
|
|||||
| Gene Name |
CD44
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Ovarian cancer [ICD-11: 2C73] | |||||
| 2 | Retinopathy [ICD-11: 9B71] | |||||
| 3 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Participates thereby in a wide variety of cellular functions including the activation, recirculation and homing of T-lymphocytes, hematopoiesis, inflammation and response to bacterial infection. Engages, through its ectodomain, extracellular matrix components such as hyaluronan/HA, collagen, growth factors, cytokines or proteases and serves as a platform for signal transduction by assembling, via its cytoplasmic domain, protein complexes containing receptor kinases and membrane proteases. Such effectors include PKN2, the RhoGTPases RAC1 and RHOA, Rho-kinases and phospholipase C that coordinate signaling pathways promoting calcium mobilization and actin-mediated cytoskeleton reorganization essential for cell migration and adhesion. Cell-surface receptor that plays a role in cell-cell interactions, cell adhesion and migration, helping them to sense and respond to changes in the tissue microenvironment.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MDKFWWHAAWGLCLVPLSLAQIDLNITCRFAGVFHVEKNGRYSISRTEAADLCKAFNSTL
PTMAQMEKALSIGFETCRYGFIEGHVVIPRIHPNSICAANNTGVYILTSNTSQYDTYCFN ASAPPEEDCTSVTDLPNAFDGPITITIVNRDGTRYVQKGEYRTNPEDIYPSNPTDDDVSS GSSSERSSTSGGYIFYTFSTVHPIPDEDSPWITDSTDRIPATTLMSTSATATETATKRQE TWDWFSWLFLPSESKNHLHTTTQMAGTSSNTISAGWEPNEENEDERDRHLSFSGSGIDDD EDFISSTISTTPRAFDHTKQNQDWTQWNPSHSNPEVLLQTTTRMTDVDRNGTTAYEGNWN PEAHPPLIHHEHHEEEETPHSTSTIQATPSSTTEETATQKEQWFGNRWHEGYRQTPKEDS HSTTGTAAASAHTSHPMQGRTTPSPEDSSWTDFFNPISHPMGRGHQAGRRMDMDSSHSIT LQPTANPNTGLVEDLDRTGPLSMTTQQSNSQSFSTSHEGLEEDKDHPTTSTLTSSNRNDV TGGRRDPNHSEGSTTLLEGYTSHYPHTKESRTFIPVTSAKTGSFGVTAVTVGDSNSNVNR SLSGDQDTFHPSGGSHTTHGSESDGHSHGSQEGGANTTSGPIRTPQIPEWLIILASLLAL ALILAVCIAVNSRRRCGQKKKLVINSGNGAVEDRKPSGLNGEASKSQEMVHLVNKESSET PDQFMTADETRNLQNVDMKIGV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T06JXV | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | A-6 | Drug Info | Phase 2 | Macular degeneration | [3] | |
| 2 | BIWA 4 | Drug Info | Phase 1 | Solid tumour/cancer | [4] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | Bivatuzumab mertansine | Drug Info | Terminated | Solid tumour/cancer | [5] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | A-6 | Drug Info | [1], [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| ECM-receptor interaction | hsa04512 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Hematopoietic cell lineage | hsa04640 | Affiliated Target |
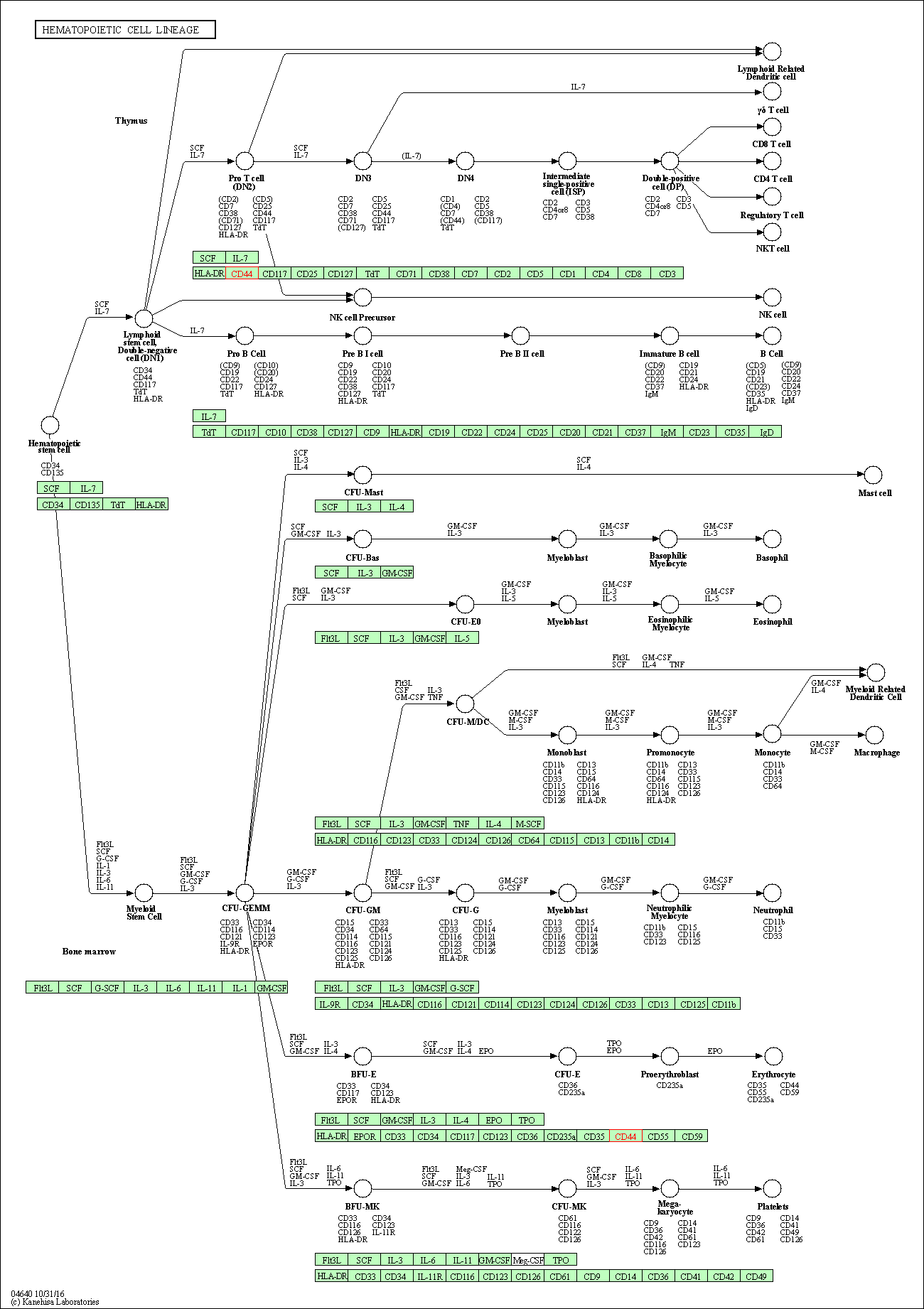
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 48 | Degree centrality | 5.16E-03 | Betweenness centrality | 6.72E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.53E-01 | Radiality | 1.44E+01 | Clustering coefficient | 5.59E-02 |
| Neighborhood connectivity | 3.19E+01 | Topological coefficient | 3.75E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 6 KEGG Pathways | + | ||||
| 1 | ECM-receptor interaction | |||||
| 2 | Hematopoietic cell lineage | |||||
| 3 | Shigellosis | |||||
| 4 | Epstein-Barr virus infection | |||||
| 5 | Proteoglycans in cancer | |||||
| 6 | MicroRNAs in cancer | |||||
| NetPath Pathway | [+] 3 NetPath Pathways | + | ||||
| 1 | IL5 Signaling Pathway | |||||
| 2 | RANKL Signaling Pathway | |||||
| 3 | TNFalpha Signaling Pathway | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Alzheimer disease-presenilin pathway | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | Osteopontin-mediated events | |||||
| 2 | a4b7 Integrin signaling | |||||
| Reactome | [+] 5 Reactome Pathways | + | ||||
| 1 | Degradation of the extracellular matrix | |||||
| 2 | Cell surface interactions at the vascular wall | |||||
| 3 | Integrin cell surface interactions | |||||
| 4 | Hyaluronan uptake and degradation | |||||
| 5 | Interferon gamma signaling | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Senescence and Autophagy in Cancer | |||||
| 2 | Wnt Signaling Pathway and Pluripotency | |||||
| 3 | Glycosaminoglycan metabolism | |||||
| 4 | Extracellular matrix organization | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Modulation of CD44 Activity by A6-Peptide.Front Immunol.2015 Mar 30;6:135. | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800011940) | |||||
| REF 4 | ClinicalTrials.gov (NCT02204046) Biodistribution Study With 186 Re-labelled Humanised Monoclonal Antibody BIWA 4 in Patients With Adenocarcinoma of the Breast. U.S. National Institutes of Health. | |||||
| REF 5 | A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin Cancer Res. 2006 Oct 15;12(20 Pt 1):6064-72. | |||||
| REF 6 | A6 peptide activates CD44 adhesive activity, induces FAK and MEK phosphorylation, and inhibits the migration and metastasis of CD44-expressing cells.Mol Cancer Ther.2011 Nov;10(11):2072-82. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

