Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T86702
(Former ID: TTDC00105)
|
|||||
| Target Name |
Matrix metalloproteinase-3 (MMP-3)
|
|||||
| Synonyms |
Transin-1; Stromelysin-1; STMY1; SL-1; MMP-3
Click to Show/Hide
|
|||||
| Gene Name |
MMP3
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Disease | [+] 7 Target-related Diseases | + | ||||
| 1 | Corneal disease [ICD-11: 9A76-9A78] | |||||
| 2 | Hepatitis virus infection [ICD-11: 1E50-1E51] | |||||
| 3 | Myocardial infarction [ICD-11: BA41-BA43] | |||||
| 4 | Osteoarthritis [ICD-11: FA00-FA05] | |||||
| 5 | Idiopathic interstitial pneumonitis [ICD-11: CB03] | |||||
| 6 | Multiple sclerosis [ICD-11: 8A40] | |||||
| 7 | Rheumatoid arthritis [ICD-11: FA20] | |||||
| Function |
Can degrade fibronectin, laminin, gelatins of type I, III, IV, and V; collagens III, IV, X, and IX, and cartilage proteoglycans. Activates procollagenase.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.24.17
|
|||||
| Sequence |
MKSLPILLLLCVAVCSAYPLDGAARGEDTSMNLVQKYLENYYDLKKDVKQFVRRKDSGPV
VKKIREMQKFLGLEVTGKLDSDTLEVMRKPRCGVPDVGHFRTFPGIPKWRKTHLTYRIVN YTPDLPKDAVDSAVEKALKVWEEVTPLTFSRLYEGEADIMISFAVREHGDFYPFDGPGNV LAHAYAPGPGINGDAHFDDDEQWTKDTTGTNLFLVAAHEIGHSLGLFHSANTEALMYPLY HSLTDLTRFRLSQDDINGIQSLYGPPPDSPETPLVPTEPVPPEPGTPANCDPALSFDAVS TLRGEILIFKDRHFWRKSLRKLEPELHLISSFWPSLPSGVDAAYEVTSKDLVFIFKGNQF WAIRGNEVRAGYPRGIHTLGFPPTVRKIDAAISDKEKNKTYFFVEDKYWRFDEKRNSMEP GFPKQIAEDFPGIDSKIDAVFEEFGFFYFFTGSSQLEFDPNAKKVTHTLKSNSWLNC Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A00079 ; BADD_A00228 ; BADD_A00237 ; BADD_A00852 ; BADD_A01161 | |||||
| HIT2.0 ID | T20H9F | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Discontinued Drug(s) | [+] 5 Discontinued Drugs | + | ||||
| 1 | GM6001 | Drug Info | Discontinued in Phase 2 | Corneal ulcer | [2] | |
| 2 | PG-530742 | Drug Info | Discontinued in Phase 2 | Myocardial infarction | [3] | |
| 3 | RS-130830 | Drug Info | Discontinued in Phase 2 | Hepatitis C virus infection | [4] | |
| 4 | BB-1101 | Drug Info | Terminated | Multiple sclerosis | [6] | |
| 5 | RO-319790 | Drug Info | Terminated | Rheumatoid arthritis | [7] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | Batimastat | Drug Info | Preclinical | Idiopathic pulmonary fibrosis | [5] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 20 Inhibitor drugs | + | ||||
| 1 | PMID29130358-Compound-Figure10(2a) | Drug Info | [1] | |||
| 2 | GM6001 | Drug Info | [8], [9], [10] | |||
| 3 | PG-530742 | Drug Info | [3], [11] | |||
| 4 | RS-130830 | Drug Info | [12] | |||
| 5 | BB-1101 | Drug Info | [14] | |||
| 6 | L-696418 | Drug Info | [15] | |||
| 7 | RO-319790 | Drug Info | [16] | |||
| 8 | 1-Methyloxy-4-Sulfone-Benzene | Drug Info | [17] | |||
| 9 | 3-Methylpyridine | Drug Info | [17] | |||
| 10 | 8-chloro-quinoline-3-carbonitrile | Drug Info | [18] | |||
| 11 | AM-2S | Drug Info | [19] | |||
| 12 | CM-352 | Drug Info | [20] | |||
| 13 | FUTOENONE | Drug Info | [21] | |||
| 14 | Hydroxyaminovaline | Drug Info | [17] | |||
| 15 | MMI270 | Drug Info | [22] | |||
| 16 | PD-169469 | Drug Info | [23] | |||
| 17 | PKF-242-484 | Drug Info | [24] | |||
| 18 | PNU-107859 | Drug Info | [25] | |||
| 19 | RS-39066 | Drug Info | [26] | |||
| 20 | UK-356618 | Drug Info | [27] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Batimastat | Drug Info | [13] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: 1-Methyloxy-4-Sulfone-Benzene | Ligand Info | |||||
| Structure Description | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | PDB:1BM6 | ||||
| Method | Solution NMR | Resolution | N.A. | Mutation | No | [28] |
| PDB Sequence |
FRTFPGIPKW
92 RKTHLTYRIV102 NYTPDLPKDA112 VDSAVEKALK122 VWEEVTPLTF132 SRLYEGEADI 142 MISFAVREHG152 DFYPFDGPGN162 VLAHAYAPGP172 GINGDAHFDD182 DEQWTKDTTG 192 TNLFLVAAHE202 IGHSLGLFHS212 ANTEALMYPL222 YHSLTDLTRF232 RLSQDDINGI 242 QSLYGPPPDS252 PET
|
|||||
|
|
||||||
| Ligand Name: 3-Methylpyridine | Ligand Info | |||||
| Structure Description | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | PDB:1BM6 | ||||
| Method | Solution NMR | Resolution | N.A. | Mutation | No | [28] |
| PDB Sequence |
FRTFPGIPKW
92 RKTHLTYRIV102 NYTPDLPKDA112 VDSAVEKALK122 VWEEVTPLTF132 SRLYEGEADI 142 MISFAVREHG152 DFYPFDGPGN162 VLAHAYAPGP172 GINGDAHFDD182 DEQWTKDTTG 192 TNLFLVAAHE202 IGHSLGLFHS212 ANTEALMYPL222 YHSLTDLTRF232 RLSQDDINGI 242 QSLYGPPPDS252 PET
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
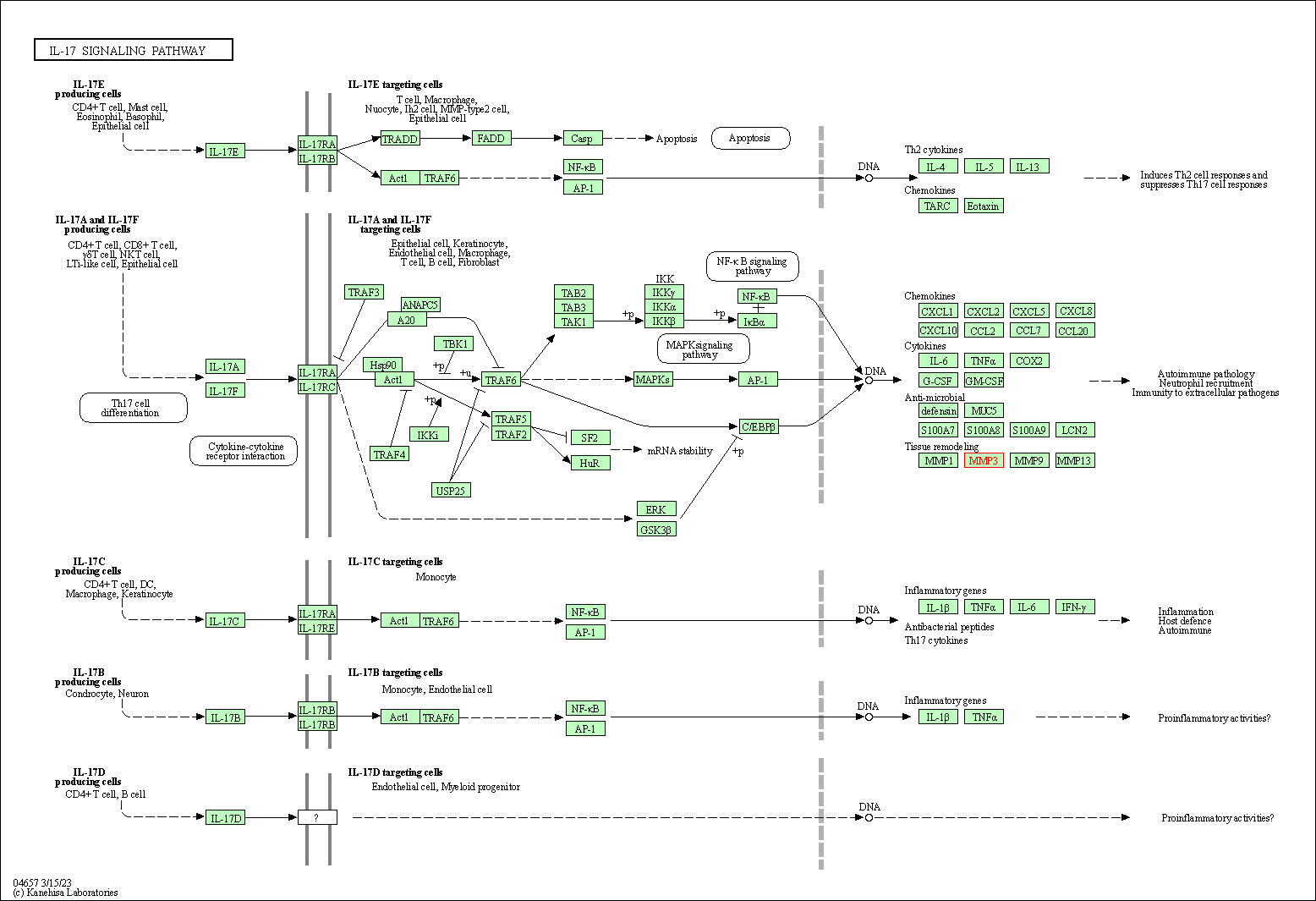
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
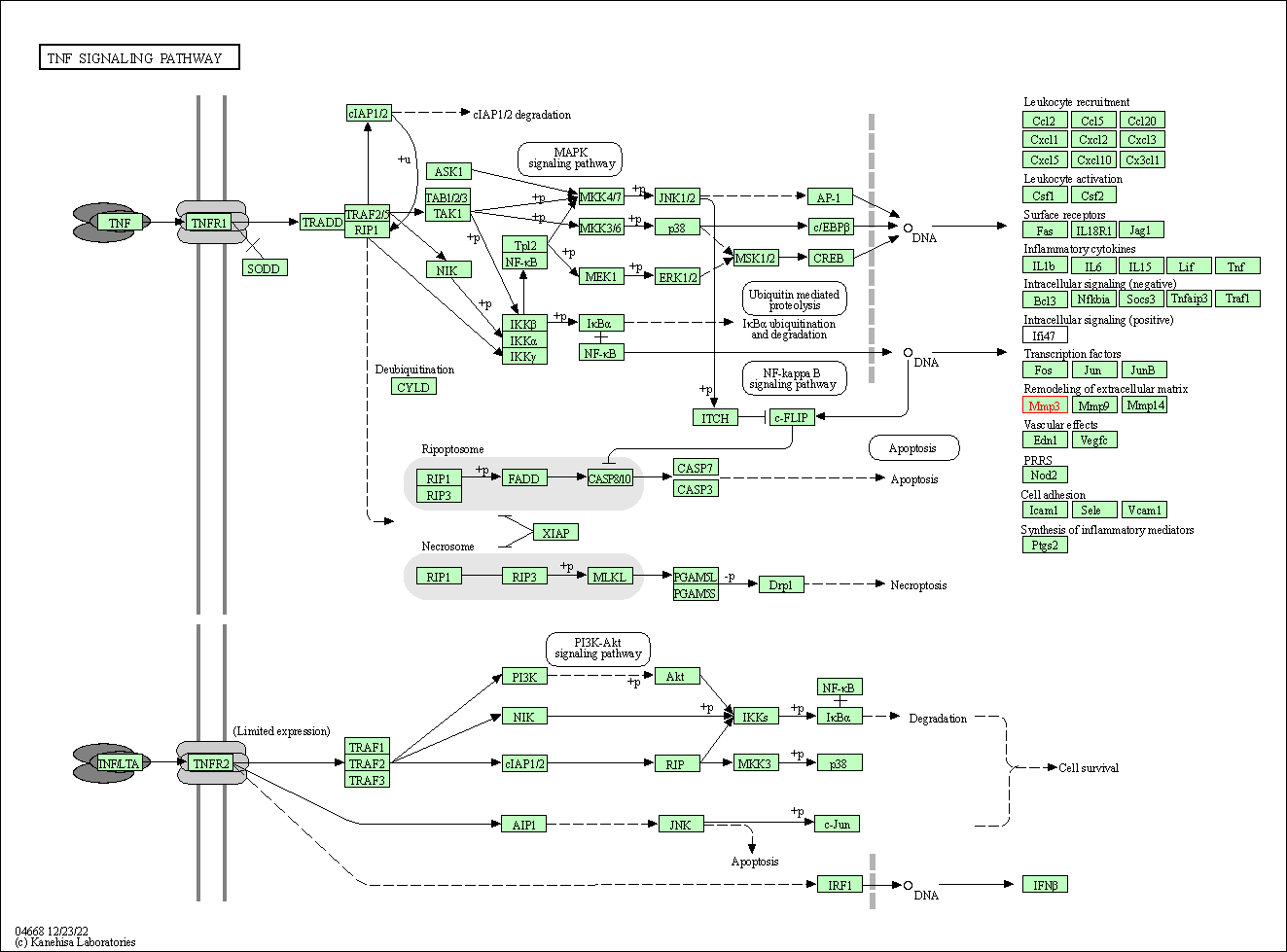
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 17 | Degree centrality | 1.83E-03 | Betweenness centrality | 9.16E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.38E-01 | Radiality | 1.42E+01 | Clustering coefficient | 3.24E-01 |
| Neighborhood connectivity | 4.62E+01 | Topological coefficient | 9.83E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | TNF signaling pathway | |||||
| 2 | Transcriptional misregulation in cancer | |||||
| 3 | Rheumatoid arthritis | |||||
| NetPath Pathway | [+] 2 NetPath Pathways | + | ||||
| 1 | IL1 Signaling Pathway | |||||
| 2 | IL4 Signaling Pathway | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Plasminogen activating cascade | |||||
| 2 | CCKR signaling map ST | |||||
| PID Pathway | [+] 3 PID Pathways | + | ||||
| 1 | Posttranslational regulation of adherens junction stability and dissassembly | |||||
| 2 | p75(NTR)-mediated signaling | |||||
| 3 | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling | |||||
| Reactome | [+] 5 Reactome Pathways | + | ||||
| 1 | Collagen degradation | |||||
| 2 | Degradation of the extracellular matrix | |||||
| 3 | Activation of Matrix Metalloproteinases | |||||
| 4 | Assembly of collagen fibrils and other multimeric structures | |||||
| 5 | EGFR Transactivation by Gastrin | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Activation of Matrix Metalloproteinases | |||||
| 2 | Gastrin-CREB signalling pathway via PKC and MAPK | |||||
| 3 | Oncostatin M Signaling Pathway | |||||
| 4 | Matrix Metalloproteinases | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| Target QSAR Model | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Gelatinase inhibitors: a patent review (2011-2017).Expert Opin Ther Pat. 2018 Jan;28(1):31-46. | |||||
| REF 2 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800001387) | |||||
| REF 3 | Selective matrix metalloproteinase inhibition attenuates progression of left ventricular dysfunction and remodeling in dogs with chronic heart fail... Am J Physiol Heart Circ Physiol. 2006 Jun;290(6):H2522-7. | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010620) | |||||
| REF 5 | Drugs and Targets in Fibrosis. Front Pharmacol. 2017 Nov 23;8:855. | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800006361) | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002350) | |||||
| REF 8 | Introduction of the 4-(4-bromophenyl)benzenesulfonyl group to hydrazide analogs of Ilomastat leads to potent gelatinase B (MMP-9) inhibitors with i... Bioorg Med Chem. 2008 Sep 15;16(18):8745-59. | |||||
| REF 9 | Blockade of tumor necrosis factor-alpha-converting enzyme improves experimental small intestinal damage by decreasing matrix metalloproteinase-3 production in rats. Scand J Gastroenterol. 2006 Nov;41(11):1320-9. | |||||
| REF 10 | Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol Cell Biol. 2008 Apr;28(7):2391-413. | |||||
| REF 11 | Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006 Mar;6(3):227-39. | |||||
| REF 12 | Structure-based design of potent and selective inhibitors of collagenase-3 (MMP-13). Bioorg Med Chem Lett. 2005 Feb 15;15(4):1101-6. | |||||
| REF 13 | Matrix metalloproteinase inhibitor BB-94 (batimastat) inhibits human colon tumor growth and spread in a patient-like orthotopic model in nude mice. Cancer Res. 1994 Sep 1;54(17):4726-8. | |||||
| REF 14 | Broad spectrum matrix metalloproteinase inhibitors: an examination of succinamide hydroxamate inhibitors with P1 C alpha gem-disubstitution. Bioorg Med Chem Lett. 1998 Jun 16;8(12):1443-8. | |||||
| REF 15 | Inhibition of matrix metalloproteinases by N-carboxyalkyl peptides containing extended alkyl residues At P1', Bioorg. Med. Chem. Lett. 5(6):539-542 (1995). | |||||
| REF 16 | The asymmetric synthesis and in vitro characterization of succinyl mercaptoalcohol and mercaptoketone inhibitors of matrix metalloproteinases. Bioorg Med Chem Lett. 1998 May 19;8(10):1163-8. | |||||
| REF 17 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 18 | Pharmacologic inhibition of tpl2 blocks inflammatory responses in primary human monocytes, synoviocytes, and blood. J Biol Chem. 2007 Nov 16;282(46):33295-304. | |||||
| REF 19 | Synthesis of hydroxypyrone- and hydroxythiopyrone-based matrix metalloproteinase inhibitors: developing a structure-activity relationship. Bioorg Med Chem Lett. 2009 Apr 1;19(7):1970-6. | |||||
| REF 20 | Discovery and safety profiling of a potent preclinical candidate, (4-[4-[[(3R)-3-(hydroxycarbamoyl)-8-azaspiro[4.5]decan-3-yl]sulfonyl]phenoxy]-N-methylbenzamide) (CM-352), for the prevention and treatment of hemorrhage. J Med Chem. 2015 Apr 9;58(7):2941-57. | |||||
| REF 21 | Inhibition of metalloproteinase by futoenone derivatives, Bioorg. Med. Chem. Lett. 5(15):1637-1642 (1995). | |||||
| REF 22 | Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002 Sep;2(9):657-72. | |||||
| REF 23 | Structural insight into the stereoselective inhibition of MMP-8 by enantiomeric sulfonamide phosphonates. J Med Chem. 2006 Feb 9;49(3):923-31. | |||||
| REF 24 | A cassette-dosing approach for improvement of oral bioavailability of dual TACE/MMP inhibitors. Bioorg Med Chem Lett. 2006 May 15;16(10):2632-6. | |||||
| REF 25 | A molecular basis for the selectivity of thiadiazole urea inhibitors with stromelysin-1 and gelatinase-A from generalized born molecular dynamics s... J Med Chem. 2004 Jun 3;47(12):3065-74. | |||||
| REF 26 | Design, synthesis, activity, and structure of a novel class of matrix metalloproteinase inhibitors containing a heterocyclic P2 P3 Bioorg. Med. Chem. Lett. 6(13):1541-1542 (1996). | |||||
| REF 27 | A potent, selective inhibitor of matrix metalloproteinase-3 for the topical treatment of chronic dermal ulcers. J Med Chem. 2003 Jul 31;46(16):3514-25. | |||||
| REF 28 | Solution structure of the catalytic domain of human stromelysin-1 complexed to a potent, nonpeptidic inhibitor. Biochemistry. 1998 Oct 6;37(40):14048-56. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

