Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T92777
(Former ID: TTDS00291)
|
|||||
| Target Name |
Intestinal maltase-glucoamylase (MGAM)
|
|||||
| Synonyms |
MGAM
Click to Show/Hide
|
|||||
| Gene Name |
MGAM
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Acute diabete complication [ICD-11: 5A2Y] | |||||
| 2 | Inborn carbohydrate metabolism error [ICD-11: 5C51] | |||||
| Function |
May serve as an alternate pathway for starch digestion when luminal alpha-amylase activity is reduced because of immaturity or malnutrition. May play a unique role in the digestion of malted dietary oligosaccharides used in food manufacturing.
Click to Show/Hide
|
|||||
| BioChemical Class |
Glycosylase
|
|||||
| UniProt ID | ||||||
| Sequence |
MARKKLKKFTTLEIVLSVLLLVLFIISIVLIVLLAKESLKSTAPDPGTTGTPDPGTTGTP
DPGTTGTTHARTTGPPDPGTTGTTPVSAECPVVNELERINCIPDQPPTKATCDQRGCCWN PQGAVSVPWCYYSKNHSYHVEGNLVNTNAGFTARLKNLPSSPVFGSNVDNVLLTAEYQTS NRFHFKLTDQTNNRFEVPHEHVQSFSGNAAASLTYQVEISRQPFSIKVTRRSNNRVLFDS SIGPLLFADQFLQLSTRLPSTNVYGLGEHVHQQYRHDMNWKTWPIFNRDTTPNGNGTNLY GAQTFFLCLEDASGLSFGVFLMNSNAMEVVLQPAPAITYRTIGGILDFYVFLGNTPEQVV QEYLELIGRPALPSYWALGFHLSRYEYGTLDNMREVVERNRAAQLPYDVQHADIDYMDER RDFTYDSVDFKGFPEFVNELHNNGQKLVIIVDPAISNNSSSSKPYGPYDRGSDMKIWVNS SDGVTPLIGEVWPGQTVFPDYTNPNCAVWWTKEFELFHNQVEFDGIWIDMNEVSNFVDGS VSGCSTNNLNNPPFTPRILDGYLFCKTLCMDAVQHWGKQYDIHNLYGYSMAVATAEAAKT VFPNKRSFILTRSTFAGSGKFAAHWLGDNTATWDDLRWSIPGVLEFNLFGIPMVGPDICG FALDTPEELCRRWMQLGAFYPFSRNHNGQGYKDQDPASFGADSLLLNSSRHYLNIRYTLL PYLYTLFFRAHSRGDTVARPLLHEFYEDNSTWDVHQQFLWGPGLLITPVLDEGAEKVMAY VPDAVWYDYETGSQVRWRKQKVEMELPGDKIGLHLRGGYIFPTQQPNTTTLASRKNPLGL IIALDENKEAKGELFWDNGETKDTVANKVYLLCEFSVTQNRLEVNISQSTYKDPNNLAFN EIKILGTEEPSNVTVKHNGVPSQTSPTVTYDSNLKVAIITDIDLLLGEAYTVEWSIKIRD EEKIDCYPDENGASAENCTARGCIWEASNSSGVPFCYFVNDLYSVSDVQYNSHGATADIS LKSSVYANAFPSTPVNPLRLDVTYHKNEMLQFKIYDPNKNRYEVPVPLNIPSMPSSTPEG QLYDVLIKKNPFGIEIRRKSTGTIIWDSQLLGFTFSDMFIRISTRLPSKYLYGFGETEHR SYRRDLEWHTWGMFSRDQPPGYKKNSYGVHPYYMGLEEDGSAHGVLLLNSNAMDVTFQPL PALTYRTTGGVLDFYVFLGPTPELVTQQYTELIGRPVMVPYWSLGFQLCRYGYQNDSEIA SLYDEMVAAQIPYDVQYSDIDYMERQLDFTLSPKFAGFPALINRMKADGMRVILILDPAI SGNETQPYPAFTRGVEDDVFIKYPNDGDIVWGKVWPDFPDVVVNGSLDWDSQVELYRAYV AFPDFFRNSTAKWWKREIEELYNNPQNPERSLKFDGMWIDMNEPSSFVNGAVSPGCRDAS LNHPPYMPHLESRDRGLSSKTLCMESQQILPDGSLVQHYNVHNLYGWSQTRPTYEAVQEV TGQRGVVITRSTFPSSGRWAGHWLGDNTAAWDQLKKSIIGMMEFSLFGISYTGADICGFF QDAEYEMCVRWMQLGAFYPFSRNHNTIGTRRQDPVSWDVAFVNISRTVLQTRYTLLPYLY TLMHKAHTEGVTVVRPLLHEFVSDQVTWDIDSQFLLGPAFLVSPVLERNARNVTAYFPRA RWYDYYTGVDINARGEWKTLPAPLDHINLHVRGGYILPWQEPALNTHLSRQKFMGFKIAL DDEGTAGGWLFWDDGQSIDTYGKGLYYLASFSASQNTMQSHIIFNNYITGTNPLKLGYIE IWGVGSVPVTSVSISVSGMVITPSFNNDPTTQVLSIDVTDRNISLHNFTSLTWISTL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T09GFP | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 4 Approved Drugs | + | ||||
| 1 | Acarbose | Drug Info | Approved | Diabetic complication | [2], [3] | |
| 2 | Miglitol | Drug Info | Approved | Diabetic complication | [3], [4] | |
| 3 | Rh-alphaglucosidase | Drug Info | Approved | Pompe disease | [5] | |
| 4 | Voglibose | Drug Info | Approved | Diabetic complication | [3] | |
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | Deoxynojirimycin | Drug Info | Phase 3 | Pompe disease | [7] | |
| 2 | PAZ-320 | Drug Info | Phase 2 | Type-2 diabetes | [8] | |
| 3 | SC-49483 | Drug Info | Phase 2 | Acquired immune deficiency syndrome | [9] | |
| 4 | Celgosivir | Drug Info | Phase 1/2 | Dengue fever | [10] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | A-76202M | Drug Info | Terminated | Acquired immune deficiency syndrome | [11] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 6 Modulator drugs | + | ||||
| 1 | Acarbose | Drug Info | [12] | |||
| 2 | Rh-alphaglucosidase | Drug Info | [14] | |||
| 3 | PAZ-320 | Drug Info | [16] | |||
| 4 | Celgosivir | Drug Info | [18] | |||
| 5 | A-76202M | Drug Info | [11] | |||
| 6 | Imino sugars | Drug Info | [25] | |||
| Inhibitor | [+] 14 Inhibitor drugs | + | ||||
| 1 | Miglitol | Drug Info | [13] | |||
| 2 | Voglibose | Drug Info | [1] | |||
| 3 | Deoxynojirimycin | Drug Info | [15] | |||
| 4 | SC-49483 | Drug Info | [17] | |||
| 5 | Maltose | Drug Info | [19] | |||
| 6 | SALACINOL | Drug Info | [20] | |||
| 7 | 1,4-dideoxy-1,4-imino-D-arabinito | Drug Info | [21] | |||
| 8 | 2,5-Dideoxy-2,5-imino-D-mannitol | Drug Info | [21] | |||
| 9 | 2-Cyclopentylaminomethyl-pyrrolidine-3,4-diol | Drug Info | [22] | |||
| 10 | 4'-(p-toluenesulfonamide)-3,4-dihydroxy chalcone | Drug Info | [23] | |||
| 11 | Double Oxidized Cysteine | Drug Info | [24] | |||
| 12 | KOTALANOL | Drug Info | [20] | |||
| 13 | Nicotinamide-Adenine-Dinucleotide | Drug Info | [24] | |||
| 14 | PONKORANOL | Drug Info | [26] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Miglitol | Ligand Info | |||||
| Structure Description | Crystal complex of N-terminal Human Maltase-Glucoamylase with miglitol | PDB:3L4W | ||||
| Method | X-ray diffraction | Resolution | 2.00 Å | Mutation | Yes | [27] |
| PDB Sequence |
VNELERINCI
16 PDQPPTKATC26 DQRGCCWNPQ36 GAVSVPWCYY46 SKNHSYHVEG56 NLVNTNAGFT 66 ARLKNLPSSP76 VFGSNVDNVL86 LTAEYQTSNR96 FHFKLTDQTN106 NRFEVPHEHV 116 QSFSGNAAAS126 LTYQVEISRQ136 PFSIKVTRRS146 NNRVLFDSSI156 GPLLFADQFL 166 QLSTRLPSTN176 VYGLGEHVHQ186 QYRHDMNWKT196 WPIFNRDTTP206 NGNGTNLYGA 216 QTFFLCLEDA226 SGLSFGVFLM236 NSNAMEVVLQ246 PAPAITYRTI256 GGILDFYVFL 266 GNTPEQVVQE276 YLELIGRPAL286 PSYWALGFHL296 SRYEYGTLDN306 MREVVERNRA 316 AQLPYDVQHA326 DIDYMDERRD336 FTYDSVDFKG346 FPEFVNELHN356 NGQKLVIIVD 366 PAISNNSSSS376 KPYGPYDRGS386 DMKIWVNSSD396 GVTPLIGEVW406 PGQTVFPDYT 416 NPNCAVWWTK426 EFELFHNQVE436 FDGIWIDMNE446 VSNFVDGSVS456 GCSTNNLNNP 466 PFTPRILDGY476 LFCKTLCMDA486 VQHWGKQYDI496 HNLYGYSMAV506 ATAEAAKTVF 516 PNKRSFILTR526 STFAGSGKFA536 AHWLGDNTAT546 WDDLRWSIPG556 VLEFNLFGIP 566 MVGPDICGFA576 LDTPEELCRR586 WMQLGAFYPF596 SRNHNGQGYK606 DQDPASFGAD 616 SLLLNSSRHY626 LNIRYTLLPY636 LYTLFFRAHS646 RGDTVARPLL656 HEFYEDNSTW 666 DVHQQFLWGP676 GLLITPVLDE686 GAEKVMAYVP696 DAVWYDYETG706 SQVRWRKQKV 716 EMELPGDKIG726 LHLRGGYIFP736 TQQPNTTTLA746 SRKNPLGLII756 ALDENKEAKG 766 ELFWDDGETK776 DTVANKVYLL786 CEFSVTQNRL796 EVNISQSTYK806 DPNNLAFNEI 816 KILGTEEPSN826 VTVKHNGVPS836 TSPTVTYDSN847 LKVAIITDID857 LLLGEAYTVE 867 WAH
|
|||||
|
|
||||||
| Ligand Name: Kotalanol | Ligand Info | |||||
| Structure Description | Crystal complex of N-terminal Human Maltase-Glucoamylase with kotalanol | PDB:3L4V | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | Yes | [27] |
| PDB Sequence |
VNELERINCI
16 PDQPPTKATC26 DQRGCCWNPQ36 GAVSVPWCYY46 SKNHSYHVEG56 NLVNTNAGFT 66 ARLKNLPSSP76 VFGSNVDNVL86 LTAEYQTSNR96 FHFKLTDQTN106 NRFEVPHEHV 116 QSFSGNAAAS126 LTYQVEISRQ136 PFSIKVTRRS146 NNRVLFDSSI156 GPLLFADQFL 166 QLSTRLPSTN176 VYGLGEHVHQ186 QYRHDMNWKT196 WPIFNRDTTP206 NGNGTNLYGA 216 QTFFLCLEDA226 SGLSFGVFLM236 NSNAMEVVLQ246 PAPAITYRTI256 GGILDFYVFL 266 GNTPEQVVQE276 YLELIGRPAL286 PSYWALGFHL296 SRYEYGTLDN306 MREVVERNRA 316 AQLPYDVQHA326 DIDYMDERRD336 FTYDSVDFKG346 FPEFVNELHN356 NGQKLVIIVD 366 PAISNNSSSS376 KPYGPYDRGS386 DMKIWVNSSD396 GVTPLIGEVW406 PGQTVFPDYT 416 NPNCAVWWTK426 EFELFHNQVE436 FDGIWIDMNE446 VSNFVDGSVS456 GCSTNNLNNP 466 PFTPRILDGY476 LFCKTLCMDA486 VQHWGKQYDI496 HNLYGYSMAV506 ATAEAAKTVF 516 PNKRSFILTR526 STFAGSGKFA536 AHWLGDNTAT546 WDDLRWSIPG556 VLEFNLFGIP 566 MVGPDICGFA576 LDTPEELCRR586 WMQLGAFYPF596 SRNHNGQGYK606 DQDPASFGAD 616 SLLLNSSRHY626 LNIRYTLLPY636 LYTLFFRAHS646 RGDTVARPLL656 HEFYEDNSTW 666 DVHQQFLWGP676 GLLITPVLDE686 GAEKVMAYVP696 DAVWYDYETG706 SQVRWRKQKV 716 EMELPGDKIG726 LHLRGGYIFP736 TQQPNTTTLA746 SRKNPLGLII756 ALDENKEAKG 766 ELFWDDGETK776 DTVANKVYLL786 CEFSVTQNRL796 EVNISQSTYK806 DPNNLAFNEI 816 KILGTEEPSN826 VTVKHNGVPS836 TSPTVTYDSN847 LKVAIITDID857 LLLGEAYTVE 867 WAH
|
|||||
|
|
ASP203
2.736
THR204
4.057
THR205
4.614
TYR299
3.526
ASP327
2.425
ILE328
3.801
ILE364
3.574
ASP366
4.936
TRP406
3.691
TRP441
3.253
ASP443
3.074
MET444
3.994
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Small proline-rich protein 3 (SPRR3) | 40.385 (21/52) | 1.93E-04 |
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Galactose metabolism | hsa00052 | Affiliated Target |
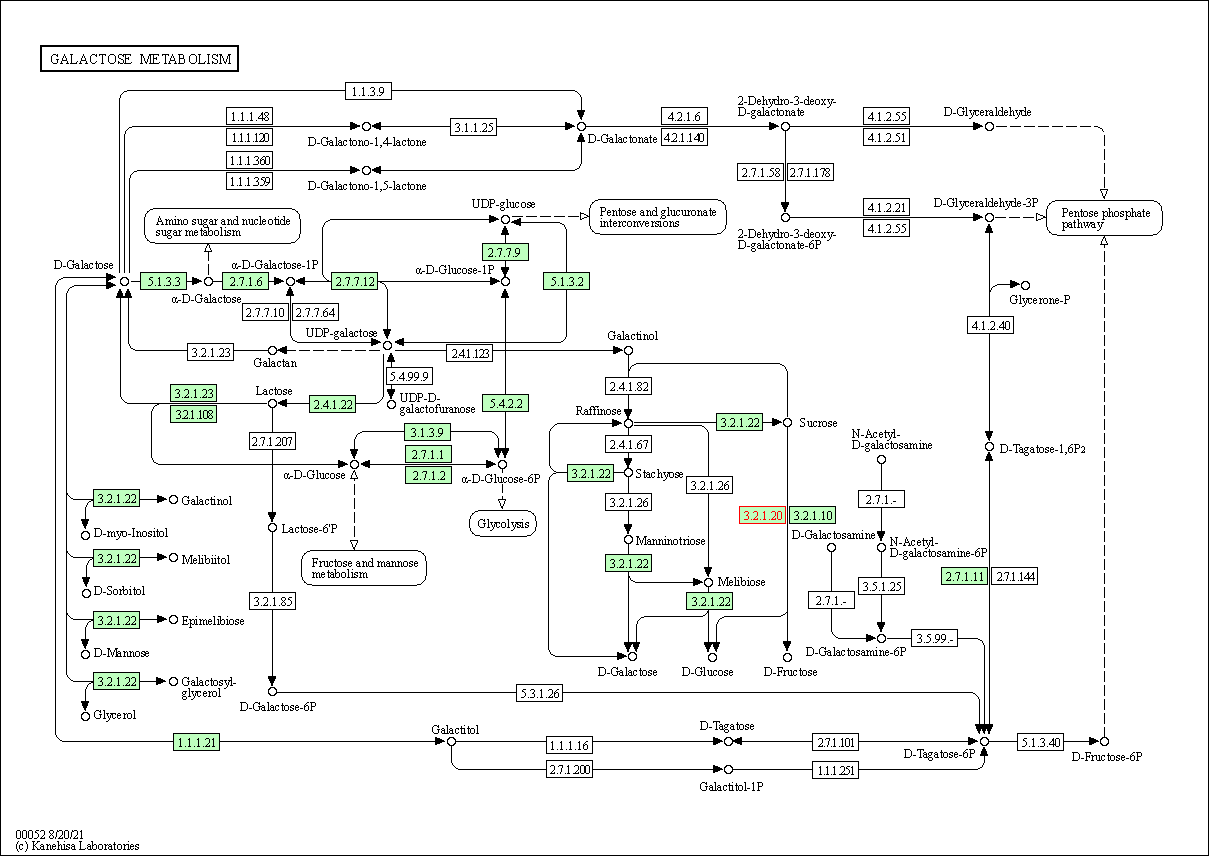
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Starch and sucrose metabolism | hsa00500 | Affiliated Target |
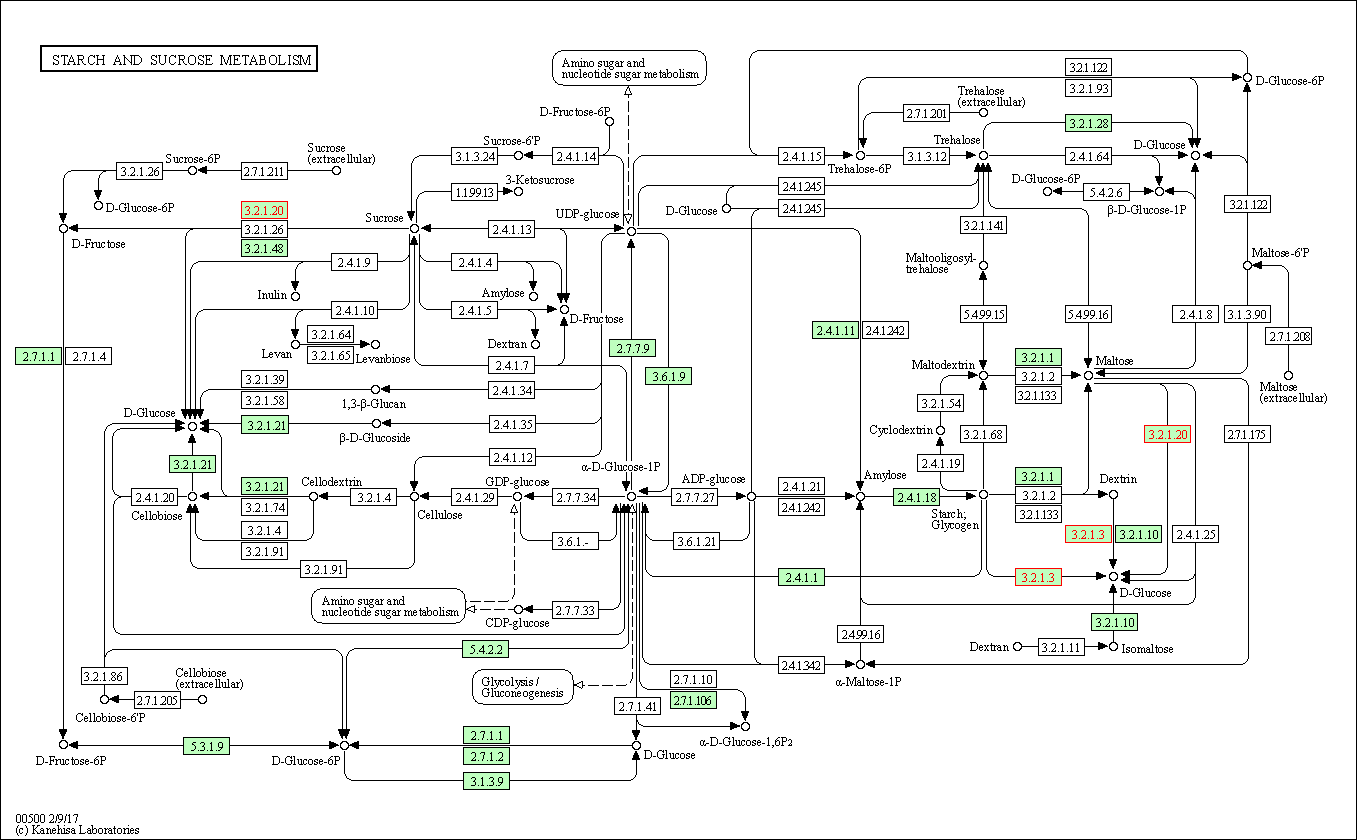
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Carbohydrate digestion and absorption | hsa04973 | Affiliated Target |
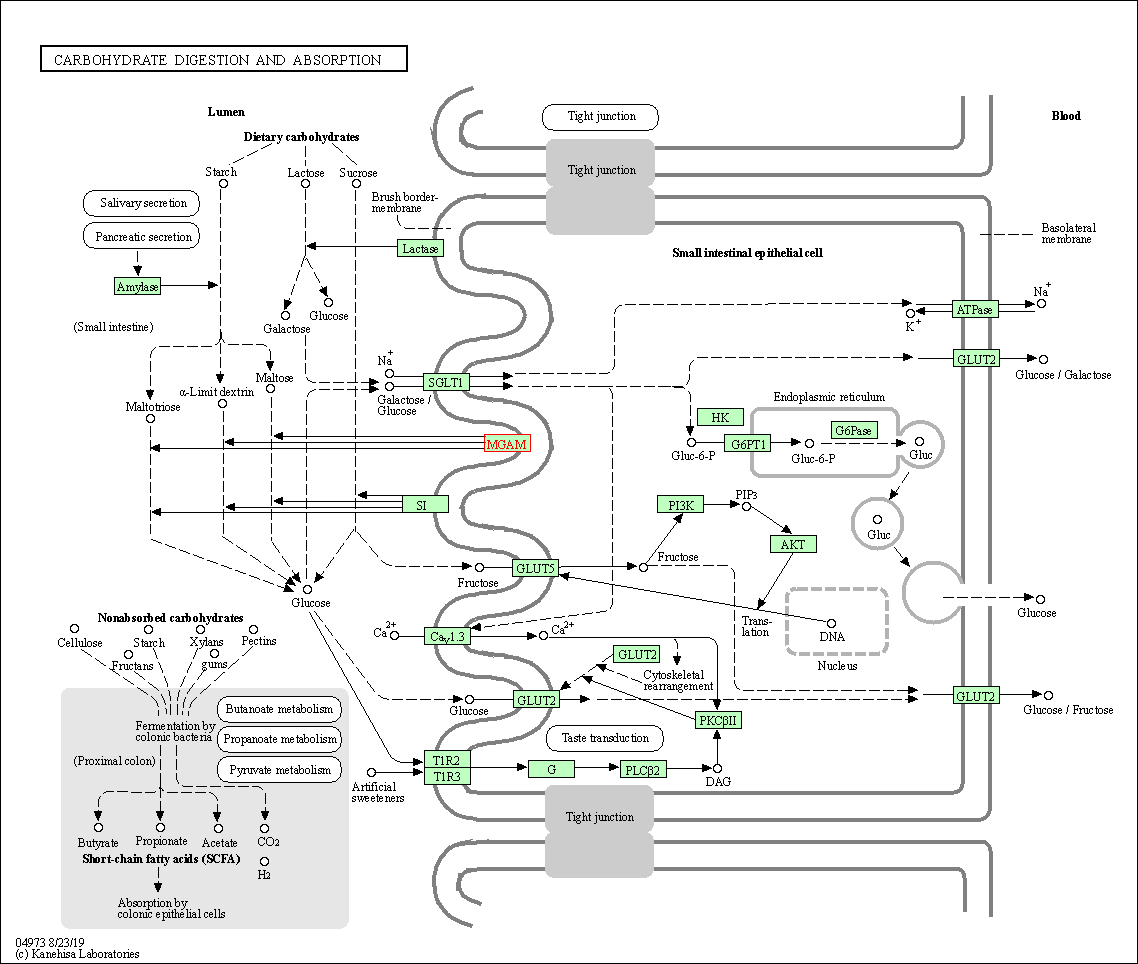
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 1.29E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 1.52E-01 | Radiality | 1.21E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 5.50E+00 | Topological coefficient | 3.00E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Galactose metabolism | |||||
| 2 | Starch and sucrose metabolism | |||||
| 3 | Metabolic pathways | |||||
| 4 | Carbohydrate digestion and absorption | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Starch and Sucrose Metabolism | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Metabolism of carbohydrates | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Effects of changeover from voglibose to acarbose on postprandial triglycerides in type 2 diabetes mellitus patients. Adv Ther. 2009 Jun;26(6):660-6. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6791). | |||||
| REF 3 | Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007 Mar;70(3):461-77. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4842). | |||||
| REF 5 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 6 | ClinicalTrials.gov (NCT01468181) A Study of LY2189265 in Japanese Participants With Type 2 Diabetes Mellitus. U.S. National Institutes of Health. | |||||
| REF 7 | The pharmacological chaperone 1-deoxynojirimycin increases the activity and lysosomal trafficking of multiple mutant forms of acid alpha-glucosidase. Hum Mutat. 2009 Dec;30(12):1683-92. | |||||
| REF 8 | ClinicalTrials.gov (NCT02060916) Study to Evaluate the Safety and Efficacy of PAZ320 in Patients With Type 2 Diabetes. U.S. National Institutes of Health. | |||||
| REF 9 | ClinicalTrials.gov (NCT00000791) A Phase II Double-Blind Study of Two Doses of SC-49483 in Combination With Zidovudine (ZDV) Versus ZDV. U.S. National Institutes of Health. | |||||
| REF 10 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800003977) | |||||
| REF 11 | Synthesis and biological activity of 4',8-dihydroxyisoflavon-7-yl D-hexopyranosides. Carbohydr Res. 2001 Oct 15;335(4):283-9. | |||||
| REF 12 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. | |||||
| REF 13 | Drug therapy of postprandial hyperglycaemia. Drugs. 1999 Jan;57(1):19-29. | |||||
| REF 14 | 2006 drug approvals: finding the niche. Nat Rev Drug Discov. 2007 Feb;6(2):99-101. | |||||
| REF 15 | Nitrogen-in-the-ring pyranoses and furanoses: structural basis of inhibition of mammalian glycosidases. J Med Chem. 1994 Oct 28;37(22):3701-6. | |||||
| REF 16 | Treatment With Novel Galactomannan Derivative Reduces 2-Hour Postprandial Glucose Excursions in Individuals With Type 2 Diabetes Treated With Oral Medications and/or Insulin. J Diabetes Sci Technol. 2014 September; 8(5): 1018-1022. | |||||
| REF 17 | Pathology of perbutylated-N-butyl-1-deoxynojiromycin (an alpha-glucosidase-1 inhibitor) in Sprague-Dawley rats. Toxicol Pathol. 1996 Sep-Oct;24(5):531-8. | |||||
| REF 18 | Celgosivir, an alpha-glucosidase I inhibitor for the potential treatment of HCV infection.Curr Opin Investig Drugs.2009 Aug;10(8):860-70. | |||||
| REF 19 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 20 | Probing the active-site requirements of human intestinal N-terminal maltase-glucoamylase: Synthesis and enzyme inhibitory activities of a six-membe... Bioorg Med Chem. 2010 Nov 15;18(22):7794-8. | |||||
| REF 21 | New sugar-mimic alkaloids from the pods of Angylocalyx pynaertii. J Nat Prod. 2002 Feb;65(2):198-202. | |||||
| REF 22 | Derivatives of (2R,3R,4S)-2-aminomethylpyrrolidine-3,4-diol are selective alpha-mannosidase inhibitors. Bioorg Med Chem Lett. 2001 Sep 17;11(18):2489-93. | |||||
| REF 23 | Sulfonamide chalcone as a new class of alpha-glucosidase inhibitors. Bioorg Med Chem Lett. 2005 Dec 15;15(24):5514-6. | |||||
| REF 24 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 25 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2627). | |||||
| REF 26 | Probing the active-site requirements of human intestinal N-terminal maltase glucoamylase: the effect of replacing the sulfate moiety by a methyl et... Bioorg Med Chem Lett. 2010 Oct 1;20(19):5686-9. | |||||
| REF 27 | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata. Biochemistry. 2010 Jan 26;49(3):443-51. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

