Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T99340
(Former ID: TTDI03510)
|
|||||
| Target Name |
Receptor-interacting protein 1 (RIPK1)
|
|||||
| Synonyms |
Receptor-interacting serine/threonine-protein kinase 1; RIP1; RIP-1; RIP; Cell death protein RIP
Click to Show/Hide
|
|||||
| Gene Name |
RIPK1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 8 Target-related Diseases | + | ||||
| 1 | Cutaneous lupus erythematosus [ICD-11: EB50-EB5Z] | |||||
| 2 | Psoriasis [ICD-11: EA90] | |||||
| 3 | Pancreatic cancer [ICD-11: 2C10] | |||||
| 4 | Alzheimer disease [ICD-11: 8A20] | |||||
| 5 | Motor neuron disease [ICD-11: 8B60] | |||||
| 6 | Nervous system paraneoplastic/autoimmune disorder [ICD-11: 8E4A] | |||||
| 7 | Rheumatoid arthritis [ICD-11: FA20] | |||||
| 8 | Ulcerative colitis [ICD-11: DD71] | |||||
| Function |
Upon activation of TNFR1 by the TNF-alpha family cytokines, TRADD and TRAF2 are recruited to the receptor. Phosphorylates DAB2IP at 'Ser-728' in a TNF-alpha-dependent manner, and thereby activates the MAP3K5-JNK apoptotic cascade. Ubiquitination by TRAF2 via 'Lys-63'-link chains acts as a critical enhancer of communication with downstream signal transducers in the mitogen-activated protein kinase pathway and the NF-kappa-B pathway, which in turn mediate downstream events including the activation of genes encoding inflammatory molecules. Polyubiquitinated protein binds to IKBKG/NEMO, the regulatory subunit of the IKK complex, a critical event for NF-kappa-B activation. Interaction with other cellular RHIM-containing adapters initiates gene activation and cell death. RIPK1 and RIPK3 association, in particular, forms a necrosis-inducing complex. Serine-threonine kinase which transduces inflammatory and cell-death signals (programmed necrosis) following death receptors ligation, activation of pathogen recognition receptors (PRRs), and DNA damage.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MQPDMSLNVIKMKSSDFLESAELDSGGFGKVSLCFHRTQGLMIMKTVYKGPNCIEHNEAL
LEEAKMMNRLRHSRVVKLLGVIIEEGKYSLVMEYMEKGNLMHVLKAEMSTPLSVKGRIIL EIIEGMCYLHGKGVIHKDLKPENILVDNDFHIKIADLGLASFKMWSKLNNEEHNELREVD GTAKKNGGTLYYMAPEHLNDVNAKPTEKSDVYSFAVVLWAIFANKEPYENAICEQQLIMC IKSGNRPDVDDITEYCPREIISLMKLCWEANPEARPTFPGIEEKFRPFYLSQLEESVEED VKSLKKEYSNENAVVKRMQSLQLDCVAVPSSRSNSATEQPGSLHSSQGLGMGPVEESWFA PSLEHPQEENEPSLQSKLQDEANYHLYGSRMDRQTKQQPRQNVAYNREEERRRRVSHDPF AQQRPYENFQNTEGKGTAYSSAASHGNAVHQPSGLTSQPQVLYQNNGLYSSHGFGTRPLD PGTAGPRVWYRPIPSHMPSLHNIPVPETNYLGNTPTMPFSSLPPTDESIKYTIYNSTGIQ IGAYNYMEIGGTSSSLLDSTNTNFKEEPAAKYQAIFDNTTSLTDKHLDPIRENLGKHWKN CARKLGFTQSQIDEIDHDYERDGLKEKVYQMLQKWVMREGIKGATVGKLAQALHQCSRID LLSSLIYVSQN Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | GSK2982772 | Drug Info | Phase 2 | Plaque psoriasis | [5] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | GSK2982772 | Drug Info | [1], [5] | |||
| 2 | Tricyclic compound 10 | Drug Info | [11] | |||
| 3 | Tricyclic compound 8 | Drug Info | [11] | |||
| 4 | Tricyclic compound 9 | Drug Info | [11] | |||
| 5 | PMID24900635C21 | Drug Info | [12] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: GSK2982772 | Ligand Info | |||||
| Structure Description | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | PDB:5TX5 | ||||
| Method | X-ray diffraction | Resolution | 2.56 Å | Mutation | Yes | [13] |
| PDB Sequence |
IKMKSSDFLE
19 SAELDSGGKV31 SLAFHRTQGL41 MIMKTVYKGP51 NCIEHNEALL61 EEAKMMNRLR 71 HSRVVKLLGV81 IIEEGKYSLV91 MEYMEKGNLM101 HVLKAEMSTP111 LSVKGRIILE 121 IIEGMAYLHG131 KGVIHKDLKP141 ENILVDNDFH151 IKIADLGLAS161 FKMWSKLNGT 189 LYYMAPEHLN199 DVNAKPTEKS209 DVYSFAVVLW219 AIFANKEPYQ235 QLIMAIKSGN 245 RPDVDDITEY255 CPREIISLMK265 LCWEANPEAR275 PTFPGIEEKF285 RPFYLSQLE |
|||||
|
|
GLY29
5.000
VAL31
3.698
ILE43
3.761
MET44
4.110
LYS45
3.646
MET67
3.632
LEU70
3.893
VAL75
3.738
VAL76
3.106
LYS77
4.551
LEU78
3.514
LEU90
3.476
|
|||||
| Ligand Name: GSK3145095 | Ligand Info | |||||
| Structure Description | Crystal structure of RIP1 kinase in complex with GSK3145095 | PDB:6RLN | ||||
| Method | X-ray diffraction | Resolution | 2.87 Å | Mutation | Yes | [14] |
| PDB Sequence |
NVIKMKSSDF
17 LEKVSLAFHR37 TQGLMIMKTV47 YKGPNCIEHN57 EALLEEAKMM67 NRLRHSRVVK 77 LLGVIIEEGK87 YSLVMEYMEK97 GNLMHVLKAE107 MSTPLSVKGR117 IILEIIEGMA 127 YLHGKGVIHK137 DLKPENILVD147 NDFHIKIADL157 GLASFKMWSK167 LNNEEHNELG 188 TLYYMAPEHL198 NDVNAKPTEK208 SDVYSFAVVL218 WAIFANKEPY228 ENAIAEQQLI 238 MAIKSGNRPD248 VDDITEYCPR258 EIISLMKLCW268 EANPEARPTF278 PGIEEKFRPF 288 YLSQLE
|
|||||
|
|
VAL31
3.404
ILE43
3.458
MET44
3.386
LYS45
3.355
MET67
4.277
LEU70
3.648
VAL75
3.642
VAL76
3.358
LYS77
4.733
LEU78
3.419
LEU90
3.042
VAL91
3.970
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
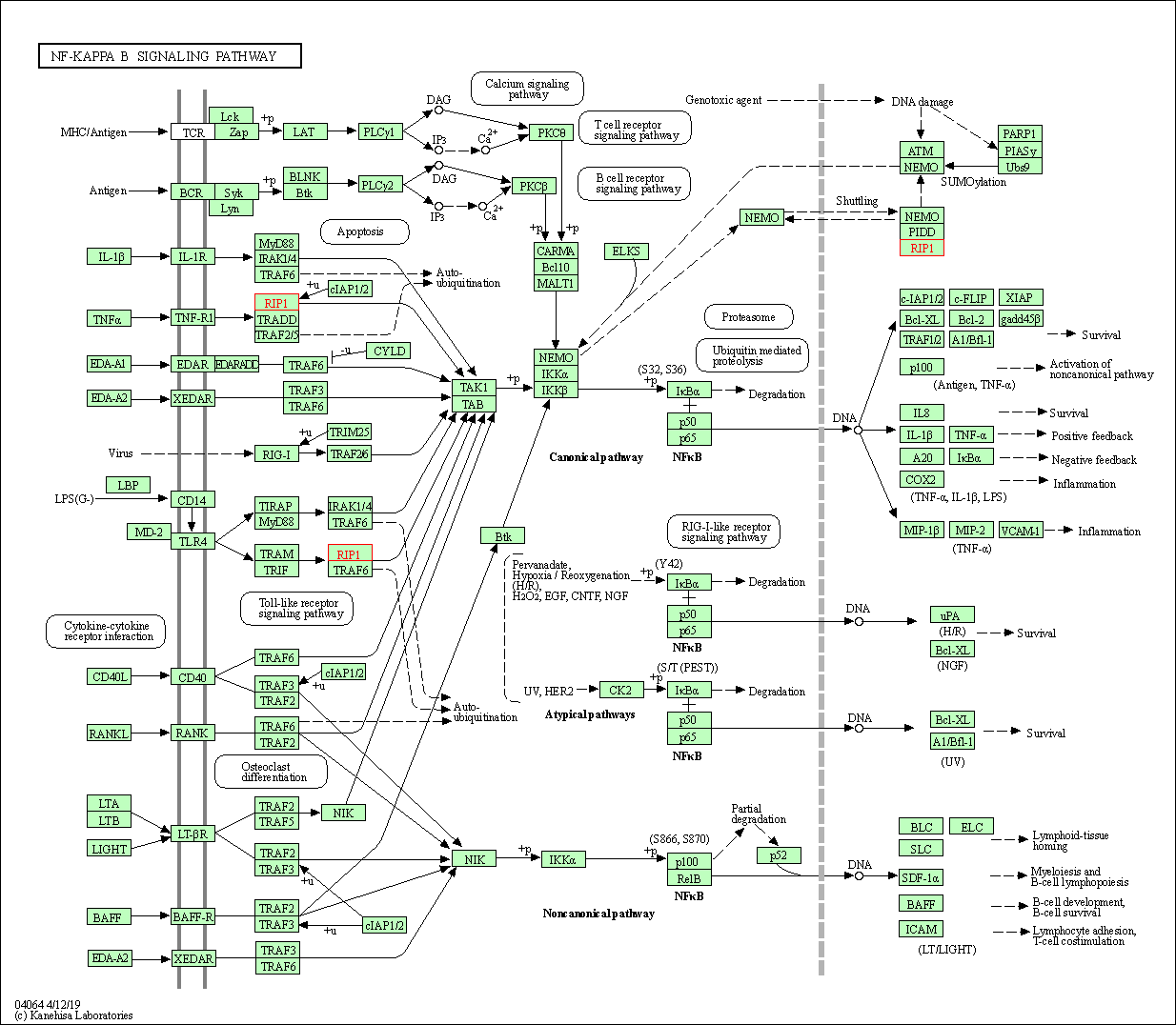
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
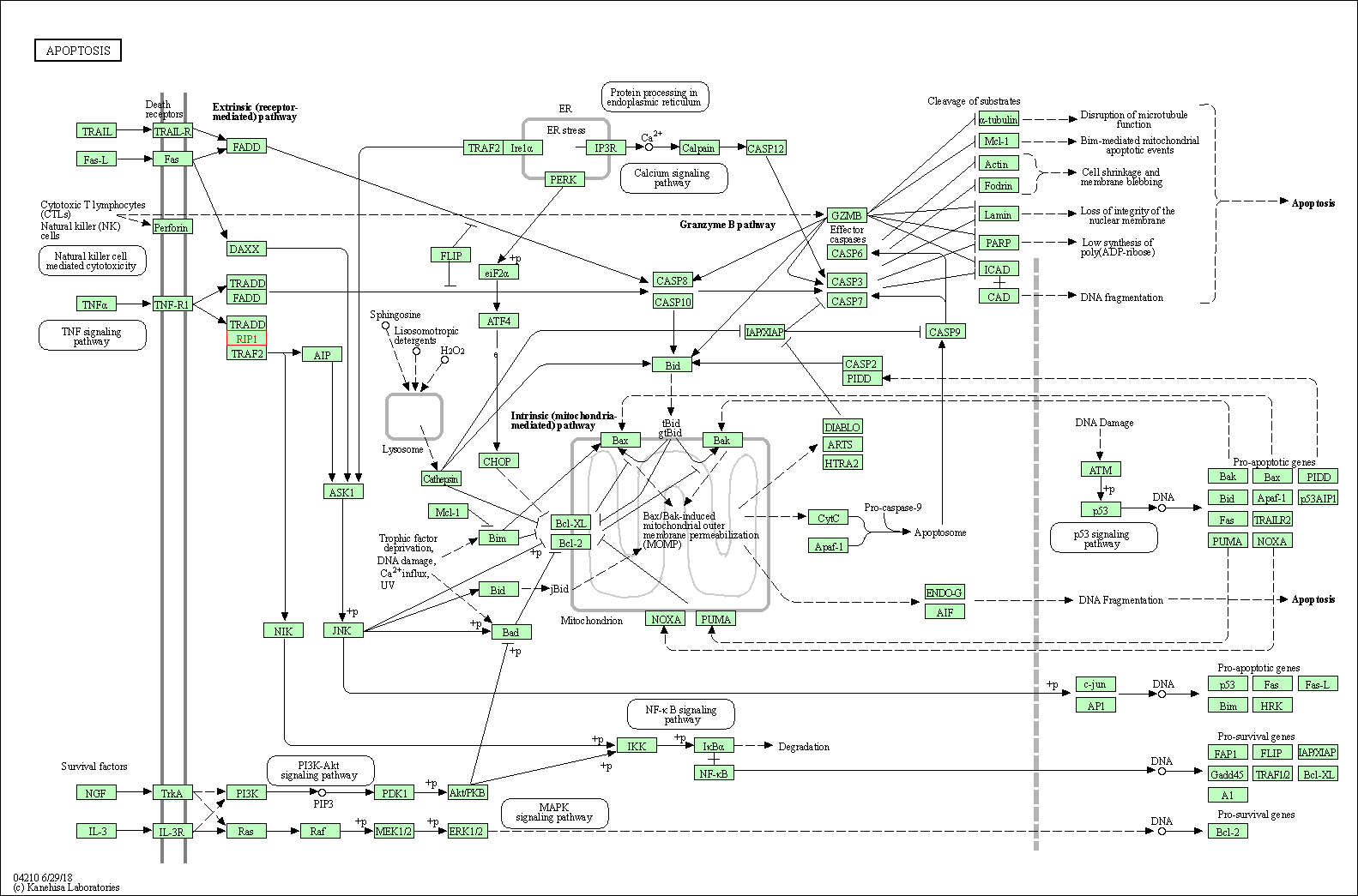
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Necroptosis | hsa04217 | Affiliated Target |
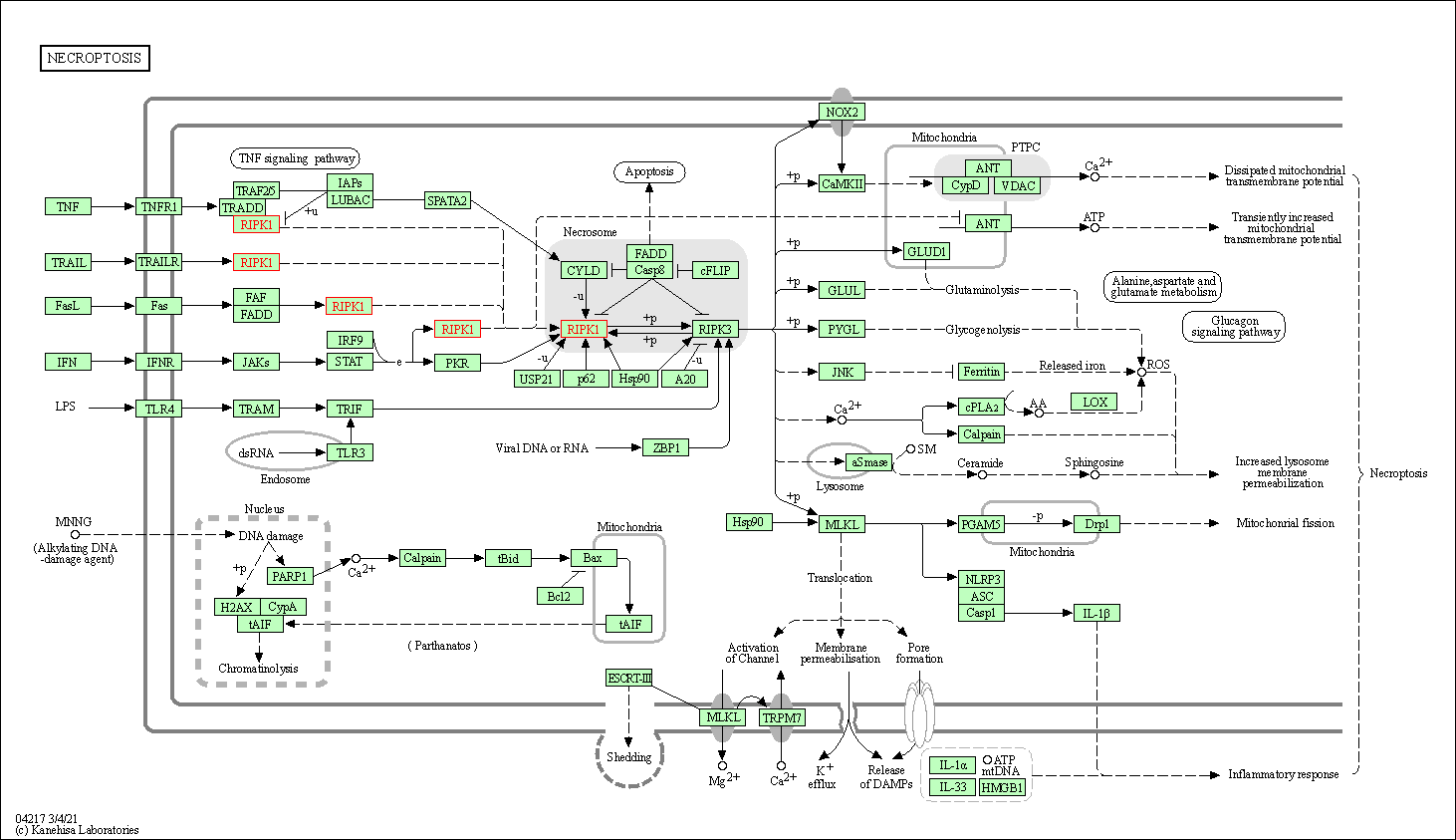
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
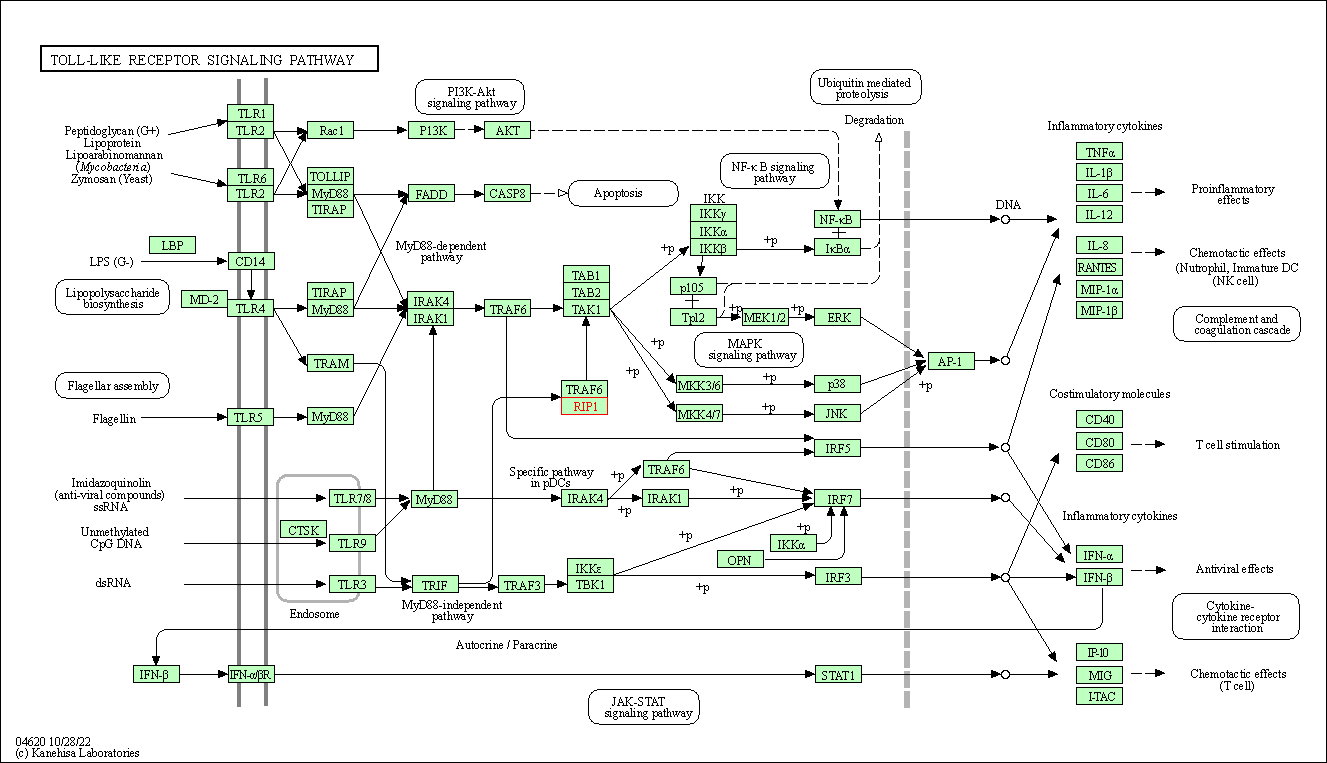
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| RIG-I-like receptor signaling pathway | hsa04622 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Cytosolic DNA-sensing pathway | hsa04623 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 52 | Degree centrality | 5.59E-03 | Betweenness centrality | 1.07E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.48E-01 | Radiality | 1.43E+01 | Clustering coefficient | 2.30E-01 |
| Neighborhood connectivity | 3.15E+01 | Topological coefficient | 5.10E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT04781816) Proof of Concept Study of SAR443122 in Patients With Cutaneous Lupus Erythematosus (CLEan). U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT05588843) A Randomized, Double-blind, Placebo Controlled, Dose-finding Study to Assess the Efficacy and Safety of SAR443122 in Adult Patients With Moderate to Severe Ulcerative Colitis. U.S.National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT05237284) A Phase 2, Multicenter, Randomized, Double-blind, Placebo-controlled Study to Evaluate the Efficacy and Safety of SAR443820 in Adult Participants With Amyotrophic Lateral Sclerosis, Followed by an Open-label Extension. U.S.National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 6 | ClinicalTrials.gov (NCT03681951) First-time-in-human (FTIH) Study of GSK3145095 Alone and in Combination With Other Anticancer Agents in Adults With Advanced Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 7 | DNL104, a Centrally Penetrant RIPK1 Inhibitor, Inhibits RIP1 Kinase Phosphorylation in a Randomized Phase I Ascending Dose Study in Healthy Volunteers. Clin Pharmacol Ther. 2020 Feb;107(2):406-414. | |||||
| REF 8 | Clinical pipeline report, company report or official report of Rigel. | |||||
| REF 9 | ClinicalTrials.gov (NCT03757325) Study to Evaluate DNL747 in Subjects With Alzheimer's Disease. U.S. National Institutes of Health. | |||||
| REF 10 | RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases. Cell Death Differ. 2020 Jan;27(1):161-175. | |||||
| REF 11 | Necroptosis inhibitors as therapeutic targets in inflammation mediated disorders - a review of the current literature and patents.Expert Opin Ther Pat. 2016 Nov;26(11):1239-1256. | |||||
| REF 12 | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis. ACS Med Chem Lett. 2013 Nov 4;4(12):1238-43. | |||||
| REF 13 | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases. J Med Chem. 2017 Feb 23;60(4):1247-1261. | |||||
| REF 14 | Identification of a RIP1 Kinase Inhibitor Clinical Candidate (GSK3145095) for the Treatment of Pancreatic Cancer. ACS Med Chem Lett. 2019 May 9;10(6):857-862. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

