Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T35486
(Former ID: TTDI01767)
|
|||||
| Target Name |
GTPase NRas (NRAS)
|
|||||
| Synonyms |
Transforming protein N-Ras; HRAS1
Click to Show/Hide
|
|||||
| Gene Name |
NRAS
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 5 Target-related Diseases | + | ||||
| 1 | Colorectal cancer [ICD-11: 2B91] | |||||
| 2 | Head and neck cancer [ICD-11: 2D42] | |||||
| 3 | Lung cancer [ICD-11: 2C25] | |||||
| 4 | Pancreatic cancer [ICD-11: 2C10] | |||||
| 5 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
Click to Show/Hide
|
|||||
| BioChemical Class |
Small GTPase
|
|||||
| UniProt ID | ||||||
| Sequence |
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAG
QEEYSAMRDQYMRTGEGFLCVFAINNSKSFADINLYREQIKRVKDSDDVPMVLVGNKCDL PTRTVDTKQAHELAKSYGIPFIETSAKTRQGVEDAFYTLVREIRQYRMKKLNSSDDGTQG CMGLPCVVM Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | GI-4000 | Drug Info | Phase 2 | Colorectal cancer | [3] | |
| 2 | Mutant ras vaccine | Drug Info | Phase 2 | Solid tumour/cancer | [4] | |
| 3 | Salirasib | Drug Info | Phase 2 | Lung cancer | [5], [6] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | Perillyl alcohol | Drug Info | Discontinued in Phase 2 | Solid tumour/cancer | [7] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Antagonist | [+] 2 Antagonist drugs | + | ||||
| 1 | Salirasib | Drug Info | [9] | |||
| 2 | KA-10X | Drug Info | [10] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Perillyl alcohol | Drug Info | [10] | |||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | APC-300 | Drug Info | [10] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Guanosine-5'-Diphosphate | Ligand Info | |||||
| Structure Description | CRYSTAL STRUCTURE OF NRAS (C118S) IN COMPLEX WITH GDP | PDB:6ZIO | ||||
| Method | X-ray diffraction | Resolution | 1.55 Å | Mutation | Yes | [11] |
| PDB Sequence |
GMTEYKLVVV
9 GAGGVGKSAL19 TIQLIQNHFV29 DEYDPTIEDS39 YRKQVVIDGE49 TCLLDILDTA 59 GQEEYSAMRD69 QYMRTGEGFL79 CVFAINNSKS89 FADINLYREQ99 IKRVKDSDDV 109 PMVLVGNKSD119 LPTRTVDTKQ129 AHELAKSYGI139 PFIETSAKTR149 QGVEDAFYTL 159 VREIRQYRMK169 K
|
|||||
|
|
ALA11
3.727
GLY12
3.859
GLY13
2.850
VAL14
3.254
GLY15
3.070
LYS16
2.775
SER17
3.046
ALA18
2.897
PHE28
3.264
VAL29
2.727
ASP30
2.698
GLU31
4.288
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Guanosine-5'-Triphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of N-ras S89D | PDB:7F68 | ||||
| Method | X-ray diffraction | Resolution | 1.24 Å | Mutation | Yes | [12] |
| PDB Sequence |
MTEYKLVVVG
10 AGGVGKSALT20 IQLIQNHFVD30 EYDPTIEDSY40 RKQVVIDGET50 CLLDILDTAG 60 REEYSAMRDQ70 YMRTGEGFLC80 VFAINNSKDF90 ADINLYREQI100 KRVKDSDDVP 110 MVLVGNKCDL120 PTRTVDTKQA130 HELAKSYGIP140 FIETSAKTRQ150 GVEDAFYTLV 160 REIRQYRMK
|
|||||
|
|
GLY10
4.657
ALA11
3.918
GLY12
3.575
GLY13
3.021
VAL14
3.247
GLY15
2.997
LYS16
2.628
SER17
2.971
ALA18
2.780
LEU19
4.978
PHE28
3.507
VAL29
3.824
ASP30
3.354
GLU31
3.126
TYR32
2.573
ASP33
4.306
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| ErbB signaling pathway | hsa04012 | Affiliated Target |
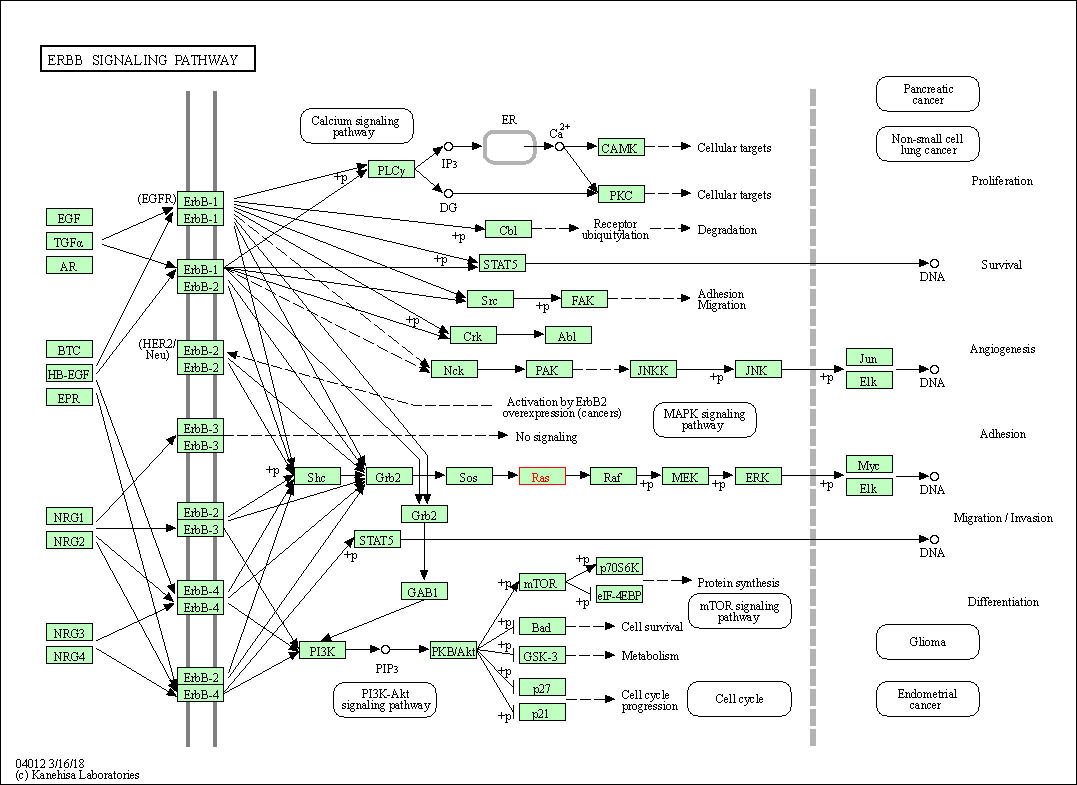
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Mitophagy - animal | hsa04137 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
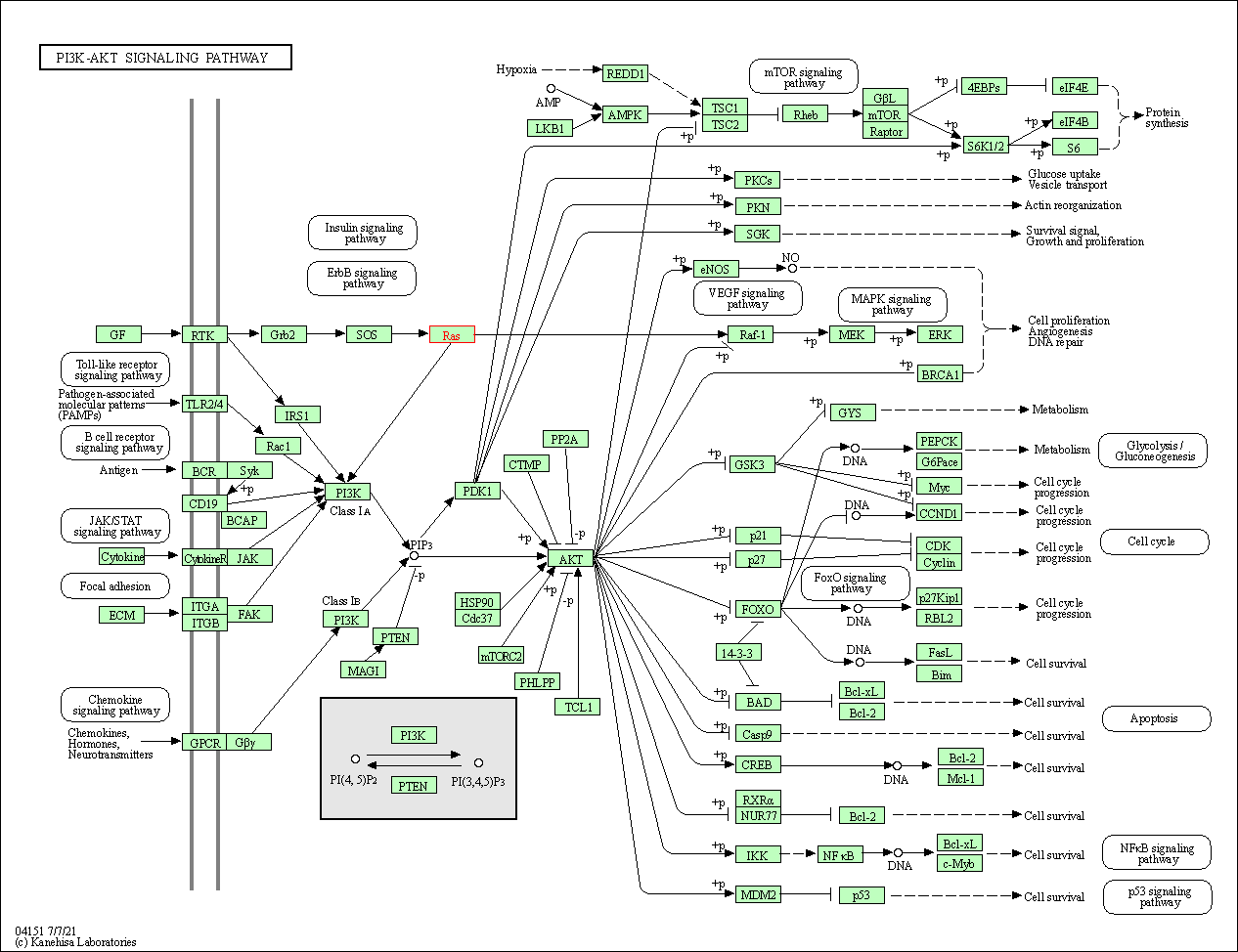
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Longevity regulating pathway | hsa04211 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Axon guidance | hsa04360 | Affiliated Target |
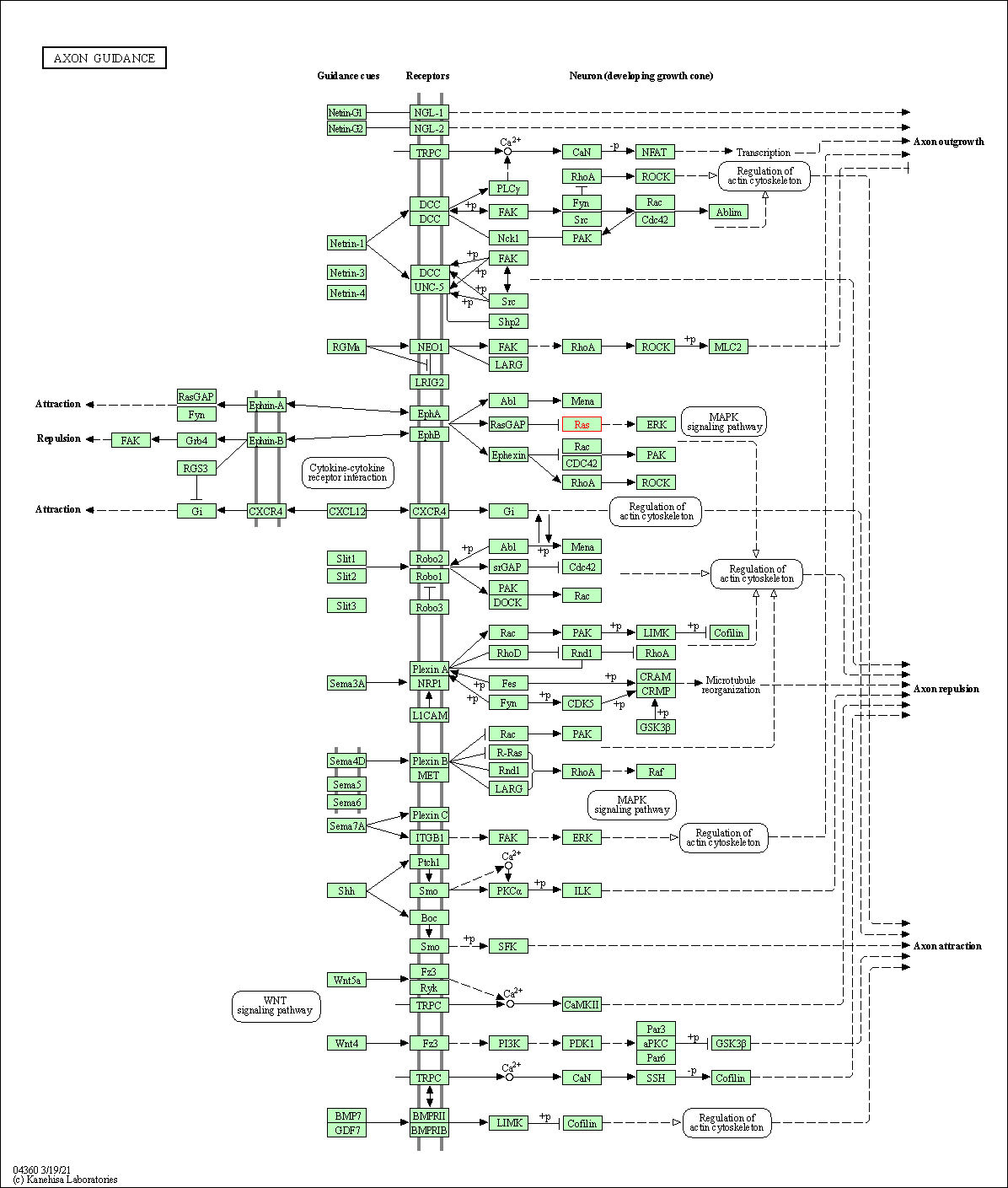
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| VEGF signaling pathway | hsa04370 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apelin signaling pathway | hsa04371 | Affiliated Target |
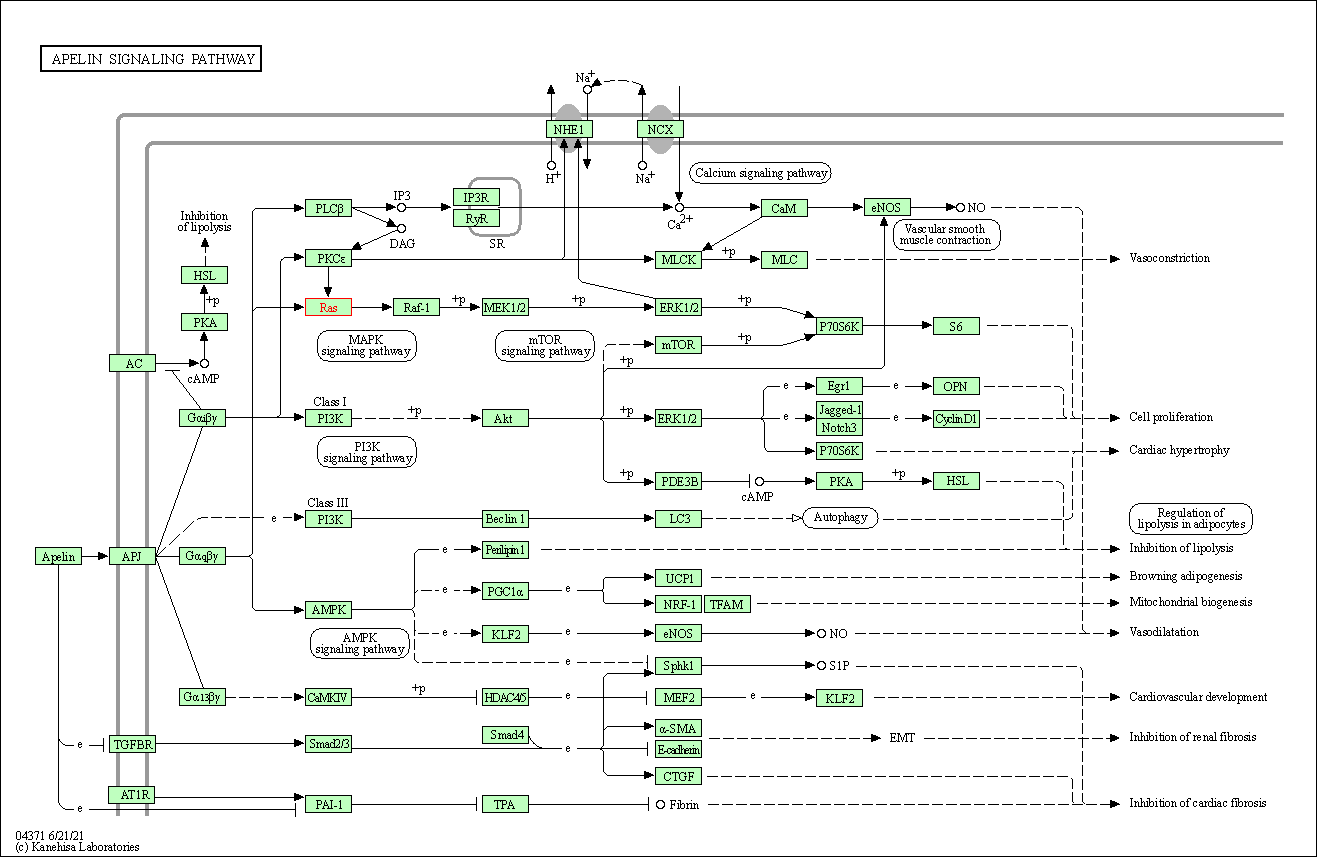
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Gap junction | hsa04540 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| B cell receptor signaling pathway | hsa04662 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Cholinergic synapse | hsa04725 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Serotonergic synapse | hsa04726 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Long-term depression | hsa04730 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Prolactin signaling pathway | hsa04917 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH secretion | hsa04929 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 43 | Degree centrality | 4.62E-03 | Betweenness centrality | 4.57E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.46E-01 | Radiality | 1.43E+01 | Clustering coefficient | 2.39E-01 |
| Neighborhood connectivity | 4.11E+01 | Topological coefficient | 6.76E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Drug Resistance Mutation (DRM) | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | The immunological and clinical effects of mutated ras peptide vaccine in combination with IL-2, GM-CSF, or both in patients with solid tumors. J Transl Med. 2014 Feb 24;12:55. | |||||
| REF 2 | ClinicalTrials.gov (NCT01199263) Paclitaxel With or Without Viral Therapy in Treating Patients With Recurrent or Persistent Ovarian Epithelial, Fallopian Tube, or Primary Peritoneal Cancer. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT00655161) A Pilot Phase 2 Trial of the Immunogenicity, and Safety of GI-4000; an Inactivated Recombinant S. Cerevisiae Expressing Mutant Ras Protein, as Consolidation Therapy Following Curative Treatment for Stage I-III Non-Small Cell Lung Cancer (NSCLC) With Tumor Sequence Confirmation of K-ras Mutation. U.S. National Institutes of Health. | |||||
| REF 4 | J Clin Oncol 30, 2012 (suppl, abstr 2577). | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6281). | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800026972) | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800004297) | |||||
| REF 8 | Phase II study of the GI-4000 KRAS vaccine after curative therapy in patients with stage I-III lung adenocarcinoma harboring a KRAS G12C, G12D, or G12V mutation. Clin Lung Cancer. 2014 Nov;15(6):405-10. | |||||
| REF 9 | The Ras inhibitor farnesylthiosalicylic acid (Salirasib) disrupts the spatiotemporal localization of active Ras: a potential treatment for cancer. Methods Enzymol. 2008;439:467-89. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2823). | |||||
| REF 11 | Drugging all RAS isoforms with one pocket. Future Med Chem. 2020 Nov;12(21):1911-1923. | |||||
| REF 12 | Crystal structure of N-ras S89D | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

