Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T46849
(Former ID: TTDR00809)
|
|||||
| Target Name |
DNA mismatch repair protein MSH2 (MSH2)
|
|||||
| Synonyms |
hMSH2; MutS protein homolog 2; Mismatch repair gene Msh2
Click to Show/Hide
|
|||||
| Gene Name |
MSH2
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Forms two different heterodimers: MutS alpha (MSH2-MSH6 heterodimer) and MutS beta (MSH2-MSH3 heterodimer) which binds to DNA mismatches thereby initiating DNA repair. When bound, heterodimers bend the DNA helix and shields approximately 20 base pairs. MutS alpha recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. MutS beta recognizes larger insertion-deletion loops up to 13 nucleotides long. After mismatch binding, MutS alpha or beta forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. Recruits DNA helicase MCM9 to chromatin which unwinds the mismatch containg DNA strand. ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. In melanocytes may modulate both UV-B-induced cell cycle regulation and apoptosis. Component of the post-replicative DNA mismatch repair system (MMR).
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MAVQPKETLQLESAAEVGFVRFFQGMPEKPTTTVRLFDRGDFYTAHGEDALLAAREVFKT
QGVIKYMGPAGAKNLQSVVLSKMNFESFVKDLLLVRQYRVEVYKNRAGNKASKENDWYLA YKASPGNLSQFEDILFGNNDMSASIGVVGVKMSAVDGQRQVGVGYVDSIQRKLGLCEFPD NDQFSNLEALLIQIGPKECVLPGGETAGDMGKLRQIIQRGGILITERKKADFSTKDIYQD LNRLLKGKKGEQMNSAVLPEMENQVAVSSLSAVIKFLELLSDDSNFGQFELTTFDFSQYM KLDIAAVRALNLFQGSVEDTTGSQSLAALLNKCKTPQGQRLVNQWIKQPLMDKNRIEERL NLVEAFVEDAELRQTLQEDLLRRFPDLNRLAKKFQRQAANLQDCYRLYQGINQLPNVIQA LEKHEGKHQKLLLAVFVTPLTDLRSDFSKFQEMIETTLDMDQVENHEFLVKPSFDPNLSE LREIMNDLEKKMQSTLISAARDLGLDPGKQIKLDSSAQFGYYFRVTCKEEKVLRNNKNFS TVDIQKNGVKFTNSKLTSLNEEYTKNKTEYEEAQDAIVKEIVNISSGYVEPMQTLNDVLA QLDAVVSFAHVSNGAPVPYVRPAILEKGQGRIILKASRHACVEVQDEIAFIPNDVYFEKD KQMFHIITGPNMGGKSTYIRQTGVIVLMAQIGCFVPCESAEVSIVDCILARVGAGDSQLK GVSTFMAEMLETASILRSATKDSLIIIDELGRGTSTYDGFGLAWAISEYIATKIGAFCMF ATHFHELTALANQIPTVNNLHVTALTTEETLTMLYQVKKGVCDQSFGIHVAELANFPKHV IECAKQKALELEEFQYIGESQGYDIMEPAAKKCYLEREQGEKIIQEFLSKVKQMPFTEMS EENITIKLKQLKAEVIAKNNSFVNEIISRIKVTT Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T76F14 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: adenosine diphosphate | Ligand Info | |||||
| Structure Description | Human MutSbeta complexed with an IDL of 3 bases (Loop3) and ADP | PDB:3THX | ||||
| Method | X-ray diffraction | Resolution | 2.70 Å | Mutation | No | [2] |
| PDB Sequence |
ESAAEVGFVR
21 FFQGMPEKPT31 TTVRLFDRGD41 FYTAHGEDAL51 LAAREVFKTQ61 GVIKYMGPAG 71 AKNLQSVVLS81 KMNFESFVKD91 LLLVRQYRVE101 VYKNRASKEN115 DWYLAYKASP 125 GNLSQFEDIL135 FIGVVGVKMS153 AVDGQRQVGV163 GYVDSIQRKL173 GLCEFPDNDQ 183 FSNLEALLIQ193 IGPKECVLPG203 GETAGDMGKL213 RQIIQRGGIL223 ITERKKADFS 233 TKDIYQDLNR243 LLKGKKGEQM253 NSAVLPEMEN263 QVAVSSLSAV273 IKFLELLSDD 283 SNFGQFELTT293 FDFSQYMKLD303 IAAVRALNLF313 QQSLAALLNK332 CKTPQGQRLV 342 NQWIKQPLMD352 KNRIEERLNL362 VEAFVEDAEL372 RQTLQEDLLR382 RFPDLNRLAK 392 KFQRQAANLQ402 DCYRLYQGIN412 QLPNVIQALE422 KHEGKHQKLL432 LAVFVTPLTD 442 LRSDFSKFQE452 MIETTLDMDQ462 VENHEFLVKP472 SFDPNLSELR482 EIMNDLEKKM 492 QSTLISAARD502 LGLDPGKQIK512 LDSSAGYYFR524 VTCKEEKVLR534 NNKNFSTVDI 544 QGVKFTNSKL556 TSLNEEYTKN566 KTEYEEAQDA576 IVKEIVNISS586 GYVEPMQTLN 596 DVLAQLDAVV606 SFAHVSNGAP616 VPYVRPAILE626 KGQGRIILKA636 SRHACVEVQI 648 AFIPNDVYFE658 KDKQMFHIIT668 GPNMGGKSTY678 IRQTGVIVLM688 AQIGCFVPCE 698 SAEVSIVDCI708 LARVGSTFMA727 EMLETASILR737 SATKDSLIII747 DELGRGTSTY 757 DGFGLAWAIS767 EYIATKIGAF777 CMFATHFHEL787 TALANQIPTV797 NNLHVTALTT 807 EETLTMLYQV817 KKGVCDQSFG827 IHVAELANFP837 KHVIECAKQK847 ALELEEFQYK 872 CYLEREQGEK882 IIQEFLSKVK892 QMPFTEMSEE902 NITIKLKQLK912 AEVIAKNNSF 922 VNEIISRI
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Mismatch repair | hsa03430 | Affiliated Target |
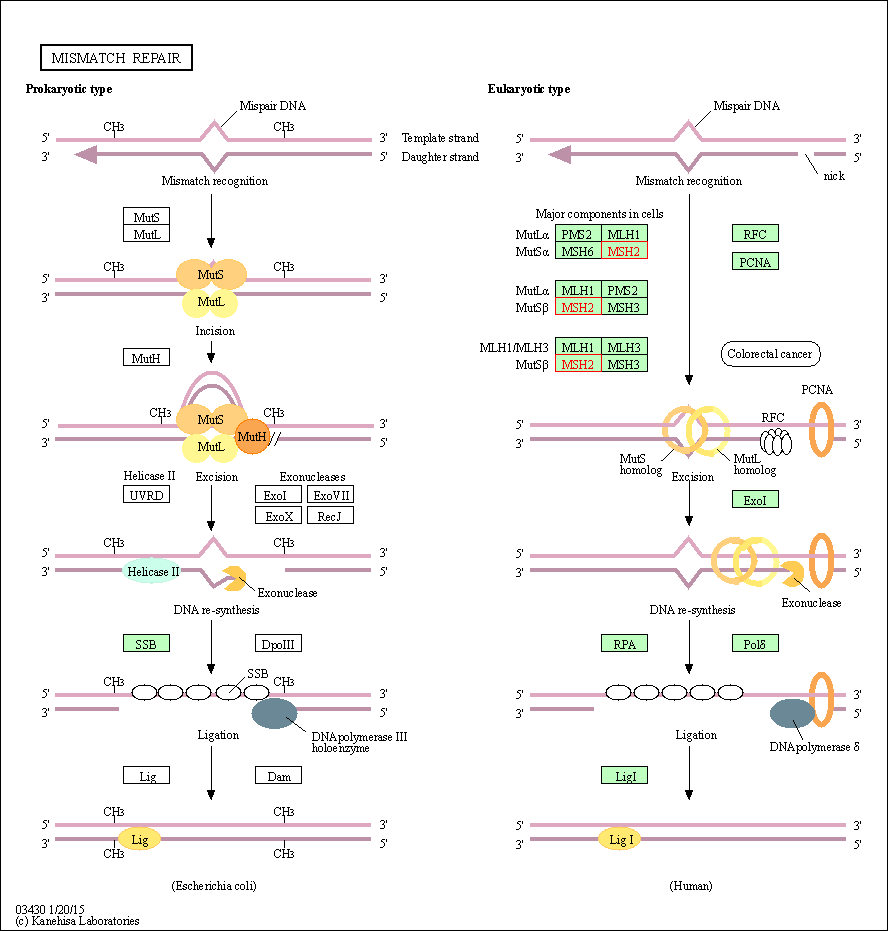
|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
| Degree | 30 | Degree centrality | 3.22E-03 | Betweenness centrality | 4.04E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.37E-01 | Radiality | 1.41E+01 | Clustering coefficient | 3.40E-01 |
| Neighborhood connectivity | 4.15E+01 | Topological coefficient | 8.29E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003 Feb 1;12(3):273-81. | |||||
| REF 2 | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nat Struct Mol Biol. 2011 Dec 18;19(1):72-8. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

