Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T55654
(Former ID: TTDNS00605)
|
|||||
| Target Name |
cAMP-dependent chloride channel (CFTR)
|
|||||
| Synonyms |
cAMPdependent chloride channel; Cystic fibrosis transmembrane conductance regulator; Channel conductancecontrolling ATPase; ATPbinding cassette subfamily C member 7; ATP-binding cassette sub-family C member 7; ABCC7
Click to Show/Hide
|
|||||
| Gene Name |
CFTR
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Cystic fibrosis [ICD-11: CA25] | |||||
| 2 | Intrathoracic organs injury [ICD-11: NB32] | |||||
| 3 | Viral intestinal infection [ICD-11: 1A2Z] | |||||
| Function |
Mediates the transport of chloride ions across the cell membrane. Channel activity is coupled to ATP hydrolysis. The ion channel is also permeable to HCO(3-); selectivity depends on the extracellular chloride concentration. Exerts its function also by modulating the activity of other ion channels and transporters. Plays an important role in airway fluid homeostasis. Contributes to the regulation of the pH and the ion content of the airway surface fluid layer and thereby plays an important role in defense against pathogens. Modulates the activity of the epithelial sodium channel (ENaC) complex, in part by regulating the cell surface expression of the ENaC complex. Inhibits the activity of the ENaC channel containing subunits SCNN1A, SCNN1B and SCNN1G. Inhibits the activity of the ENaC channel containing subunits SCNN1D, SCNN1B and SCNN1G, but not of the ENaC channel containing subunits SCNN1A, SCNN1B and SCNN1G. May regulate bicarbonate secretion and salvage in epithelial cells by regulating the transporter SLC4A7. Can inhibit the chloride channel activity of ANO1. Plays a role in the chloride and bicarbonate homeostasis during sperm epididymal maturation and capacitation. Epithelial ion channel that plays an important role in the regulation of epithelial ion and water transport and fluid homeostasis.
Click to Show/Hide
|
|||||
| BioChemical Class |
ABC transporter
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 5.6.1.6
|
|||||
| Sequence |
MQRSPLEKASVVSKLFFSWTRPILRKGYRQRLELSDIYQIPSVDSADNLSEKLEREWDRE
LASKKNPKLINALRRCFFWRFMFYGIFLYLGEVTKAVQPLLLGRIIASYDPDNKEERSIA IYLGIGLCLLFIVRTLLLHPAIFGLHHIGMQMRIAMFSLIYKKTLKLSSRVLDKISIGQL VSLLSNNLNKFDEGLALAHFVWIAPLQVALLMGLIWELLQASAFCGLGFLIVLALFQAGL GRMMMKYRDQRAGKISERLVITSEMIENIQSVKAYCWEEAMEKMIENLRQTELKLTRKAA YVRYFNSSAFFFSGFFVVFLSVLPYALIKGIILRKIFTTISFCIVLRMAVTRQFPWAVQT WYDSLGAINKIQDFLQKQEYKTLEYNLTTTEVVMENVTAFWEEGFGELFEKAKQNNNNRK TSNGDDSLFFSNFSLLGTPVLKDINFKIERGQLLAVAGSTGAGKTSLLMVIMGELEPSEG KIKHSGRISFCSQFSWIMPGTIKENIIFGVSYDEYRYRSVIKACQLEEDISKFAEKDNIV LGEGGITLSGGQRARISLARAVYKDADLYLLDSPFGYLDVLTEKEIFESCVCKLMANKTR ILVTSKMEHLKKADKILILHEGSSYFYGTFSELQNLQPDFSSKLMGCDSFDQFSAERRNS ILTETLHRFSLEGDAPVSWTETKKQSFKQTGEFGEKRKNSILNPINSIRKFSIVQKTPLQ MNGIEEDSDEPLERRLSLVPDSEQGEAILPRISVISTGPTLQARRRQSVLNLMTHSVNQG QNIHRKTTASTRKVSLAPQANLTELDIYSRRLSQETGLEISEEINEEDLKECFFDDMESI PAVTTWNTYLRYITVHKSLIFVLIWCLVIFLAEVAASLVVLWLLGNTPLQDKGNSTHSRN NSYAVIITSTSSYYVFYIYVGVADTLLAMGFFRGLPLVHTLITVSKILHHKMLHSVLQAP MSTLNTLKAGGILNRFSKDIAILDDLLPLTIFDFIQLLLIVIGAIAVVAVLQPYIFVATV PVIVAFIMLRAYFLQTSQQLKQLESEGRSPIFTHLVTSLKGLWTLRAFGRQPYFETLFHK ALNLHTANWFLYLSTLRWFQMRIEMIFVIFFIAVTFISILTTGEGEGRVGIILTLAMNIM STLQWAVNSSIDVDSLMRSVSRVFKFIDMPTEGKPTKSTKPYKNGQLSKVMIIENSHVKK DDIWPSGGQMTVKDLTAKYTEGGNAILENISFSISPGQRVGLLGRTGSGKSTLLSAFLRL LNTEGEIQIDGVSWDSITLQQWRKAFGVIPQKVFIFSGTFRKNLDPYEQWSDQEIWKVAD EVGLRSVIEQFPGKLDFVLVDGGCVLSHGHKQLMCLARSVLSKAKILLLDEPSAHLDPVT YQIIRRTLKQAFADCTVILCEHRIEAMLECQQFLVIEENKVRQYDSIQKLLNERSLFRQA ISPSDRVKLFPHRNSSKCKSKPQIAALKEETEEEVQDTRL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T53KY7 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 3 Approved Drugs | + | ||||
| 1 | Crofelemer | Drug Info | Approved | HIV-associated diarrhoea | [3], [5], [6] | |
| 2 | Lumacaftor + ivacaftor | Drug Info | Approved | Cystic fibrosis | [4] | |
| 3 | Tezacaftor and ivacaftor | Drug Info | Approved | Acute lung injury | [7] | |
| Clinical Trial Drug(s) | [+] 5 Clinical Trial Drugs | + | ||||
| 1 | ABBV-2222 | Drug Info | Phase 2 | Cystic fibrosis | [4] | |
| 2 | GLPG-1837 | Drug Info | Phase 2 | Cystic fibrosis | [4] | |
| 3 | iOWH032 | Drug Info | Phase 2 | Diarrhea | [12] | |
| 4 | ABBV-2737 | Drug Info | Phase 1 | Cystic fibrosis | [4] | |
| 5 | ABBV-3067 | Drug Info | Phase 1 | Cystic fibrosis | [4] | |
| Mode of Action | [+] 6 Modes of Action | + | ||||
| Modulator | [+] 4 Modulator drugs | + | ||||
| 1 | Crofelemer | Drug Info | [3], [6] | |||
| 2 | ABBV-2222 | Drug Info | [4] | |||
| 3 | ABBV-2737 | Drug Info | [4] | |||
| 4 | ABBV-3067 | Drug Info | [4] | |||
| Enhancer | [+] 2 Enhancer drugs | + | ||||
| 1 | Lumacaftor + ivacaftor | Drug Info | [4] | |||
| 2 | GLPG-1837 | Drug Info | [4] | |||
| Binder | [+] 1 Binder drugs | + | ||||
| 1 | Tezacaftor and ivacaftor | Drug Info | [7] | |||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | iOWH032 | Drug Info | [14] | |||
| Activator | [+] 6 Activator drugs | + | ||||
| 1 | CBIQ | Drug Info | [15] | |||
| 2 | phenylglycine-01 | Drug Info | [15] | |||
| 3 | sulfonamide-01 | Drug Info | [15] | |||
| 4 | UCCF-029 | Drug Info | [15] | |||
| 5 | UCCF-339 | Drug Info | [15] | |||
| 6 | UCCF-853 | Drug Info | [15] | |||
| Blocker (channel blocker) | [+] 2 Blocker (channel blocker) drugs | + | ||||
| 1 | CFTRinh-172 | Drug Info | [15] | |||
| 2 | GlyH-101 | Drug Info | [15] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Ivacaftor | Ligand Info | |||||
| Structure Description | The complex of phosphorylated human delta F508 cystic fibrosis transmembrane conductance regulator (CFTR) with Trikafta [elexacaftor (VX-445), tezacaftor (VX-661), ivacaftor (VX-770)] and ATP/Mg | PDB:8EIQ | ||||
| Method | Electron microscopy | Resolution | 3.00 Å | Mutation | Yes | [16] |
| PDB Sequence |
MQRSPLEKAS
10 VVSKLFFSWT20 RPILRKGYRQ30 RLELSDIYQI40 PSVDSADNLS50 EKLEREWDRE 60 LASKKNPKLI70 NALRRCFFWR80 FMFYGIFLYL90 GEVTKAVQPL100 LLGRIIASYD 110 PDNKEERSIA120 IYLGIGLCLL130 FIVRTLLLHP140 AIFGLHHIGM150 QMRIAMFSLI 160 YKKTLKLSSR170 VLDKISIGQL180 VSLLSNNLNK190 FDEGLALAHF200 VWIAPLQVAL 210 LMGLIWELLQ220 ASAFCGLGFL230 IVLALFQAGL240 GRMMMKYRDQ250 RAGKISERLV 260 ITSEMIENIQ270 SVKAYCWEEA280 MEKMIENLRQ290 TELKLTRKAA300 YVRYFNSSAF 310 FFSGFFVVFL320 SVLPYALIKG330 IILRKIFTTI340 SFCIVLRMAV350 TRQFPWAVQT 360 WYDSLGAINK370 IQDFLQKQEY380 KTLEYNLTTT390 EVVMENVTAF400 WEGTPVLKDI 444 NFKIERGQLL454 AVAGSTGAGK464 TSLLMVIMGE474 LEPSEGKIKH484 SGRISFCSQF 494 SWIMPGTIKE504 NIIGVSYDEY515 RYRSVIKACQ525 LEEDISKFAE535 KDNIVLGEGG 545 ITLSGGQRAR555 ISLARAVYKD565 ADLYLLDSPF575 GYLDVLTEKE585 IFESCVCKLM 595 ANKTRILVTS605 KMEHLKKADK615 ILILHEGSSY625 FYGTFSELQN635 LWNTYLRYIT 854 VHKSLIFVLI864 WCLVIFLAEV874 AASLVVLWLL884 GSYAVIITST910 SSYYVFYIYV 920 GVADTLLAMG930 FFRGLPLVHT940 LITVSKILHH950 KMLHSVLQAP960 MSTLNTLKAG 970 GILNRFSKDI980 AILDDLLPLT990 IFDFIQLLLI1000 VIGAIAVVAV1010 LQPYIFVATV 1020 PVIVAFIMLR1030 AYFLQTSQQL1040 KQLESEGRSP1050 IFTHLVTSLK1060 GLWTLRAFGR 1070 QPYFETLFHK1080 ALNLHTANWF1090 LYLSTLRWFQ1100 MRIEMIFVIF1110 FIAVTFISIL 1120 TTGEGEGRVG1130 IILTLAMNIM1140 STLQWAVNSS1150 IDVDSLMRSV1160 SRVFKFIDMP 1170 TEGIWPSGGQ1209 MTVKDLTAKY1219 TEGGNAILEN1229 ISFSISPGQR1239 VGLLGRTGSG 1249 KSTLLSAFLR1259 LLNTEGEIQI1269 DGVSWDSITL1279 QQWRKAFGVI1289 PQKVFIFSGT 1299 FRKNLDPYEQ1309 WSDQEIWKVA1319 DEVGLRSVIE1329 QFPGKLDFVL1339 VDGGCVLSHG 1349 HKQLMCLARS1359 VLSKAKILLL1369 DQPSAHLDPV1379 TYQIIRRTLK1389 QAFADCTVIL 1399 CEHRIEAMLE1409 CQQFLVIEEN1419 KVRQYDSIQK1429 LLNERSLFRQ1439 AISPSDRVKL 1449 FP
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer | PDB:2PZE | ||||
| Method | X-ray diffraction | Resolution | 1.70 Å | Mutation | Yes | [17] |
| PDB Sequence |
SLTTTEVVME
395 NVTAFWEEGG437 TPVLKDINFK447 IERGQLLAVA457 GSTGAGKTSL467 LMMIMGELEP 477 SEGKIKHSGR487 ISFCSQFSWI497 MPGTIKENII507 FGVSYDEYRY517 RSVIKACQLE 527 EDISKFAEKD537 NIVLGEGGIT547 LSGGQRARIS557 LARAVYKDAD567 LYLLDSPFGY 577 LDVLTEKEIF587 ESCVCKLMAN597 KTRILVTSKM607 EHLKKADKIL617 ILHEGSSYFY 627 GTFSELQNLD639 FSSKLM
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| ABC transporters | hsa02010 | Affiliated Target |
|
| Class: Environmental Information Processing => Membrane transport | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| AMPK signaling pathway | hsa04152 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Gastric acid secretion | hsa04971 | Affiliated Target |
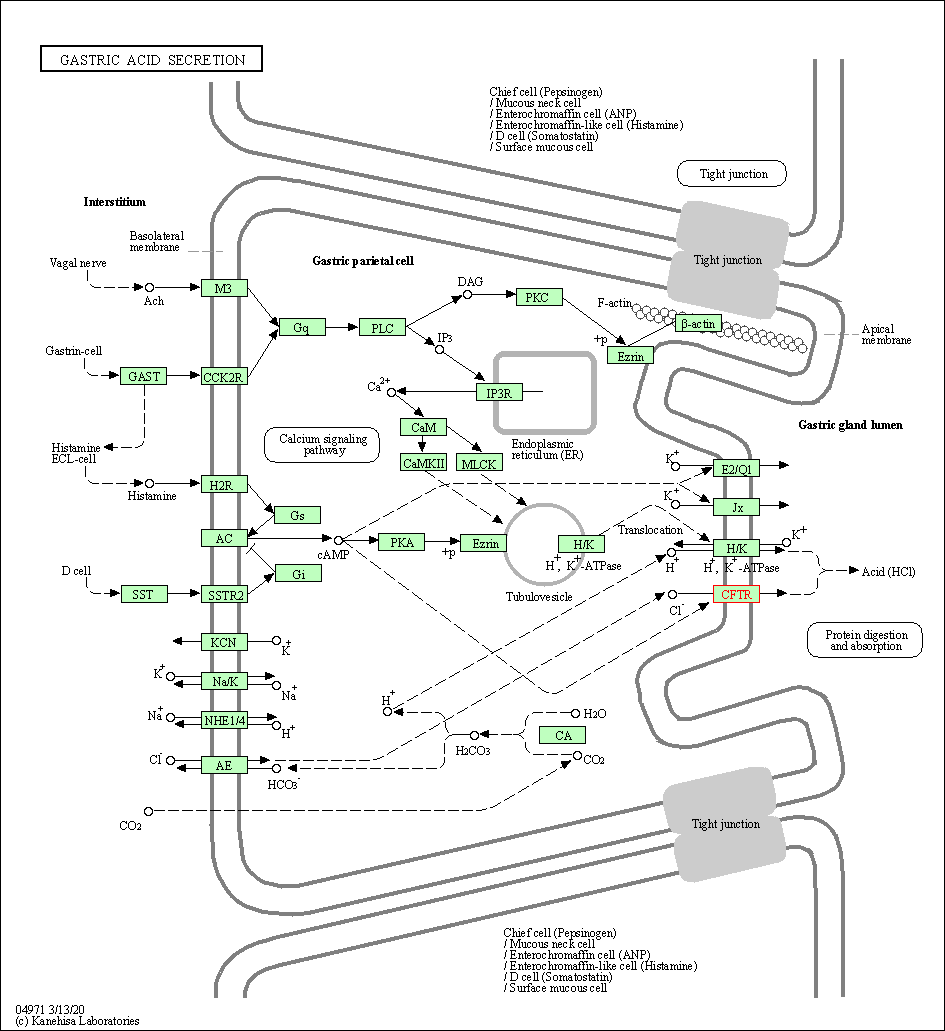
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Pancreatic secretion | hsa04972 | Affiliated Target |
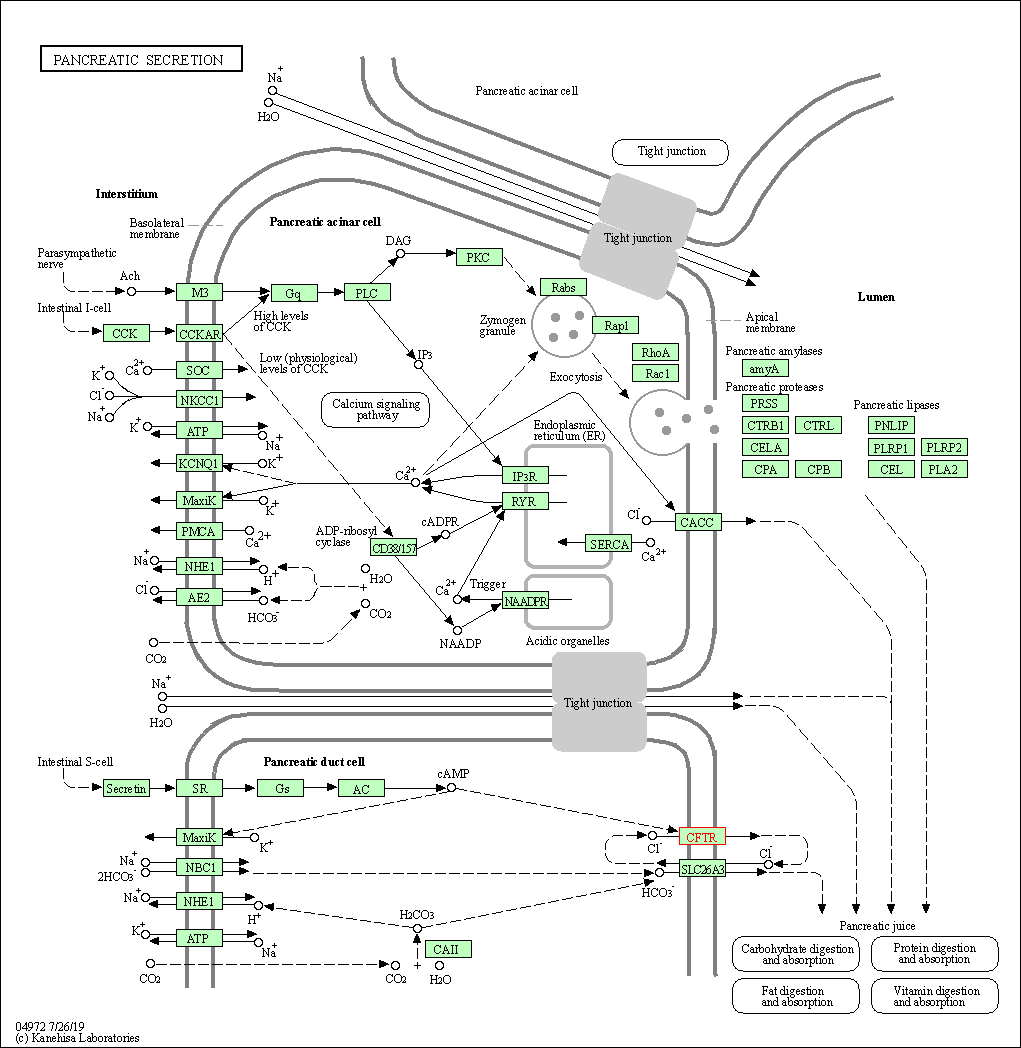
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Bile secretion | hsa04976 | Affiliated Target |
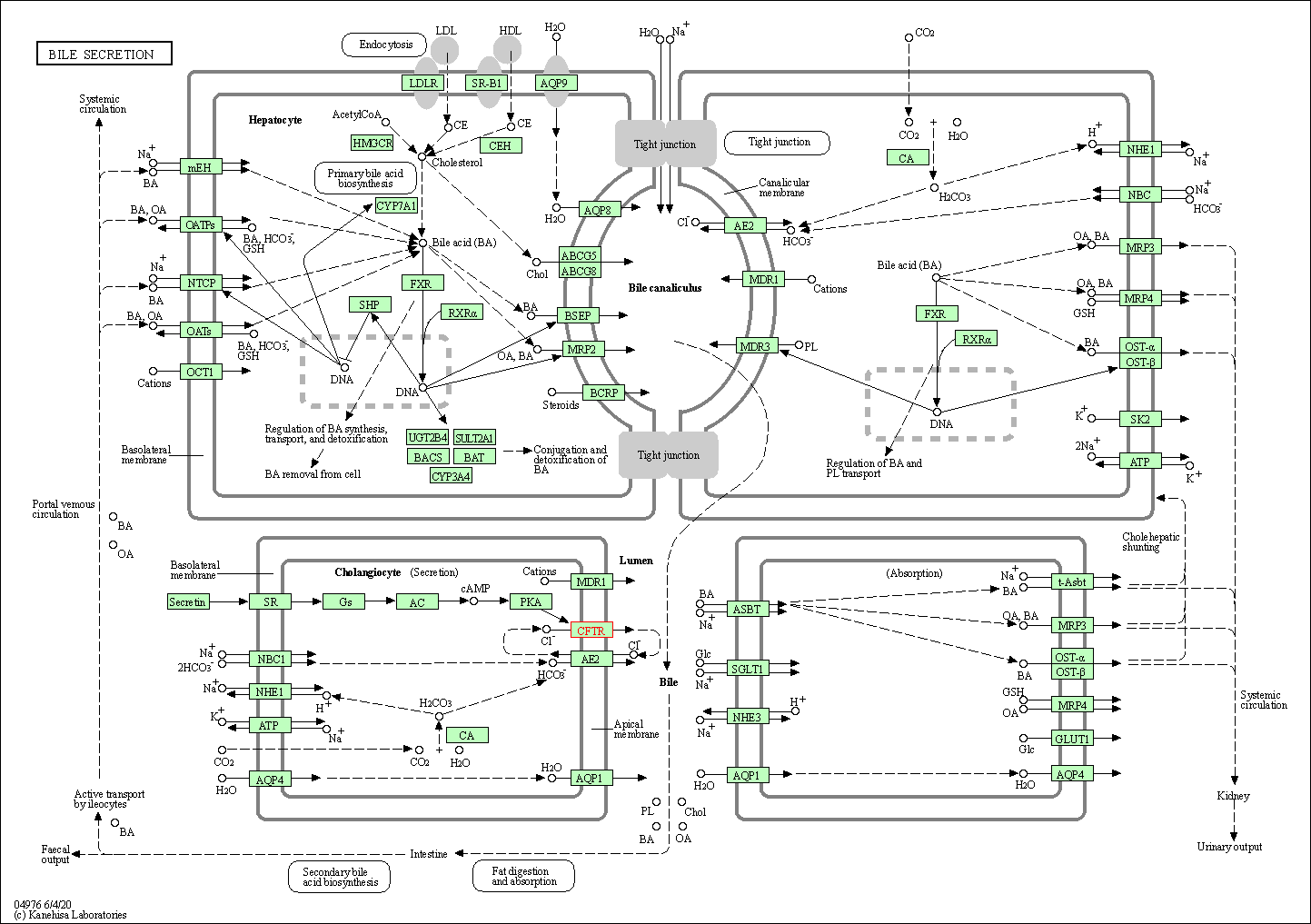
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 20 | Degree centrality | 2.15E-03 | Betweenness centrality | 4.01E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.44E-01 | Radiality | 1.43E+01 | Clustering coefficient | 7.37E-02 |
| Neighborhood connectivity | 3.02E+01 | Topological coefficient | 6.47E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 7 KEGG Pathways | + | ||||
| 1 | ABC transporters | |||||
| 2 | cAMP signaling pathway | |||||
| 3 | AMPK signaling pathway | |||||
| 4 | Gastric acid secretion | |||||
| 5 | Pancreatic secretion | |||||
| 6 | Bile secretion | |||||
| 7 | Vibrio cholerae infection | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | ABC-family proteins mediated transport | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | ABC-family proteins mediated transport | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4342). | |||||
| REF 3 | Nat Rev Drug Discov. 2013 Feb;12(2):87-90. | |||||
| REF 4 | Antibodies and venom peptides: new modalities for ion channels. Nat Rev Drug Discov. 2019 May;18(5):339-357. | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7453). | |||||
| REF 6 | Crofelemer, an antisecretory antidiarrheal proanthocyanidin oligomer extracted from Croton lechleri, targets two distinct intestinal chloride channels. Mol Pharmacol. 2010 Jan;77(1):69-78. | |||||
| REF 7 | 2018 FDA drug approvals.Nat Rev Drug Discov. 2019 Feb;18(2):85-89. | |||||
| REF 8 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 9 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7481). | |||||
| REF 10 | ClinicalTrials.gov (NCT00865904) Study of VX-809 in Cystic Fibrosis Subjects With the 508-CFTR Gene Mutation. U.S. National Institutes of Health. | |||||
| REF 11 | ClinicalTrials.gov (NCT04268823) A Randomized, Subjects and Investigator Blinded, Placebo Controlled Parallel Group Study to Assess the Mode of Action of QBW251 in Patients With Chronic Obstructive Pulmonary Disease (COPD). U.S.National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT02111304) Efficacy of iOWH032 in Dehydrating Cholera. U.S. National Institutes of Health. | |||||
| REF 13 | ClinicalTrials.gov (NCT05668741) A Phase 1 Single Dose Escalation Study Evaluating the Safety and Tolerability of VX-522 in Subjects 18 Years of Age and Older With Cystic Fibrosis and a CFTR Genotype Not Responsive to CFTR Modulator Therapy. U.S.National Institutes of Health. | |||||
| REF 14 | Developing novel antisecretory drugs to treat infectious diarrhea. Future Med Chem. 2011 Aug;3(10):1317-25. | |||||
| REF 15 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 707). | |||||
| REF 16 | Molecular structures reveal synergistic rescue of Delta508 CFTR by Trikafta modulators. Science. 2022 Oct 21;378(6617):284-290. | |||||
| REF 17 | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel. 2010 May;23(5):375-84. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

