Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T63484
(Former ID: TTDR00008)
|
|||||
| Target Name |
Glucose-6-phosphate dehydrogenase (G6PD)
|
|||||
| Synonyms |
Glucose-6-phosphate 1-dehydrogenase
Click to Show/Hide
|
|||||
| Gene Name |
G6PD
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Chronic obstructive pulmonary disease [ICD-11: CA22] | |||||
| Function |
The main function of this enzyme is to provide reducing power (NADPH) and pentose phosphates for fatty acid and nucleic acid synthesis. Catalyzes the rate-limiting step of the oxidative pentose-phosphate pathway, which represents a route for the dissimilation of carbohydrates besides glycolysis.
Click to Show/Hide
|
|||||
| BioChemical Class |
CH-OH donor oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.1.1.49
|
|||||
| Sequence |
MAEQVALSRTQVCGILREELFQGDAFHQSDTHIFIIMGASGDLAKKKIYPTIWWLFRDGL
LPENTFIVGYARSRLTVADIRKQSEPFFKATPEEKLKLEDFFARNSYVAGQYDDAASYQR LNSHMNALHLGSQANRLFYLALPPTVYEAVTKNIHESCMSQIGWNRIIVEKPFGRDLQSS DRLSNHISSLFREDQIYRIDHYLGKEMVQNLMVLRFANRIFGPIWNRDNIACVILTFKEP FGTEGRGGYFDEFGIIRDVMQNHLLQMLCLVAMEKPASTNSDDVRDEKVKVLKCISEVQA NNVVLGQYVGNPDGEGEATKGYLDDPTVPRGSTTATFAAVVLYVENERWDGVPFILRCGK ALNERKAEVRLQFHDVAGDIFHQQCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESEL DLTYGNRYKNVKLPDAYERLILDVFCGSQMHFVRSDELREAWRIFTPLLHQIELEKPKPI PYIYGSRGPTEADELMKRVGFQYEGTYKWVNPHKL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A01930 ; BADD_A01933 ; BADD_A06020 ; BADD_A06440 | |||||
| HIT2.0 ID | T87UKX | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Prasterone | Drug Info | Approved | Chronic obstructive pulmonary disease | [2] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | CBF-BS2 | Drug Info | Discontinued in Phase 2 | Rheumatoid arthritis | [3] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Prasterone | Drug Info | [1] | |||
| 2 | EPIANDROSTERONE | Drug Info | [4] | |||
| 3 | 2'-Monophosphoadenosine 5'-Diphosphoribose | Drug Info | [6] | |||
| 4 | Hydroxyacetic Acid | Drug Info | [6] | |||
| 5 | Metazamide | Drug Info | [1] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | CBF-BS2 | Drug Info | [5] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: NADP+ | Ligand Info | |||||
| Structure Description | Crystal structure of Human G6PD Canton | PDB:6JYU | ||||
| Method | X-ray diffraction | Resolution | 1.89 Å | Mutation | Yes | [7] |
| PDB Sequence |
SDTHIFIIMG
38 ASGDLAKKKI48 YPTIWWLFRD58 GLLPENTFIV68 GYARSRLTVA78 DIRKQSEPFF 88 KATPEEKLKL98 EDFFARNSYV108 AGQYDDAASY118 QRLNSHMNAL128 HLGSQANRLF 138 YLALPPTVYE148 AVTKNIHESC158 MSQIGWNRII168 VEKPFGRDLQ178 SSDRLSNHIS 188 SLFREDQIYR198 IDHYLGKEMV208 QNLMVLRFAN218 RIFGPIWNRD228 NIACVILTFK 238 EPFGTEGRGG248 YFDEFGIIRD258 VMQNHLLQML268 CLVAMEKPAS278 TNSDDVRDEK 288 VKVLKCISEV298 QANNVVLGQY308 VGNPDGEGEA318 TKGYLDDPTV328 PRGSTTATFA 338 AVVLYVENER348 WDGVPFILRC358 GKALNERKAE368 VRLQFHDVAG378 DIFHQQCKRN 388 ELVIRVQPNE398 AVYTKMMTKK408 PGMFFNPEES418 ELDLTYGNRY428 KNVKLPDAYE 438 RLILDVFCGS448 QMHFVRSDEL458 LEAWRIFTPL468 LHQIELEKPK478 PIPYIYGSRG 488 PTEADELMKR498 VGFQYEGTYK508 WVNPH
|
|||||
|
|
THR236
4.607
LYS238
2.466
ASN363
4.492
GLU364
3.574
LYS366
2.482
GLU368
4.218
ARG370
2.857
GLU389
4.139
VAL391
4.389
ARG393
2.727
ASN397
3.691
ALA399
3.894
TYR401
2.510
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: beta-D-glucose-6-phosphate | Ligand Info | |||||
| Structure Description | Structure of G6PD-D200N tetramer bound to NADP+ and G6P | PDB:7SNI | ||||
| Method | Electron microscopy | Resolution | 2.50 Å | Mutation | No | [8] |
| PDB Sequence |
QSDTHIFIIM
37 GASGDLAKKK47 IYPTIWWLFR57 DGLLPENTFI67 VGYARSRLTV77 ADIRKQSEPF 87 FKATPEEKLK97 LEDFFARNSY107 VAGQYDDAAS117 YQRLNSHMNA127 LHLGSQANRL 137 FYLALPPTVY147 EAVTKNIHES157 CMSQIGWNRI167 IVEKPFGRDL177 QSSDRLSNHI 187 SSLFREDQIY197 RINHYLGKEM207 VQNLMVLRFA217 NRIFGPIWNR227 DNIACVILTF 237 KEPFGTEGRG247 GYFDEFGIIR257 DVMQNHLLQM267 LCLVAMEKPA277 STNSDDVRDE 287 KVKVLKCISE297 VQANNVVLGQ307 YVGNPDGEGE317 ATKGYLDDPT327 VPRGSTTATF 337 AAVVLYVENE347 RWDGVPFILR357 CGKALNERKA367 EVRLQFHDVA377 GDIFHQQCKR 387 NELVIRVQPN397 EAVYTKMMTK407 KPGMFFNPEE417 SELDLTYGNR427 YKNVKLPDAY 437 ERLILDVFCG447 SQMHFVRSDE457 LREAWRIFTP467 LLHQIELEKP477 KPIPYIYGSR 487 GPTEADELMK497 RVGFQYEGTY507 KWVNPHKL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Pentose phosphate pathway | hsa00030 | Affiliated Target |
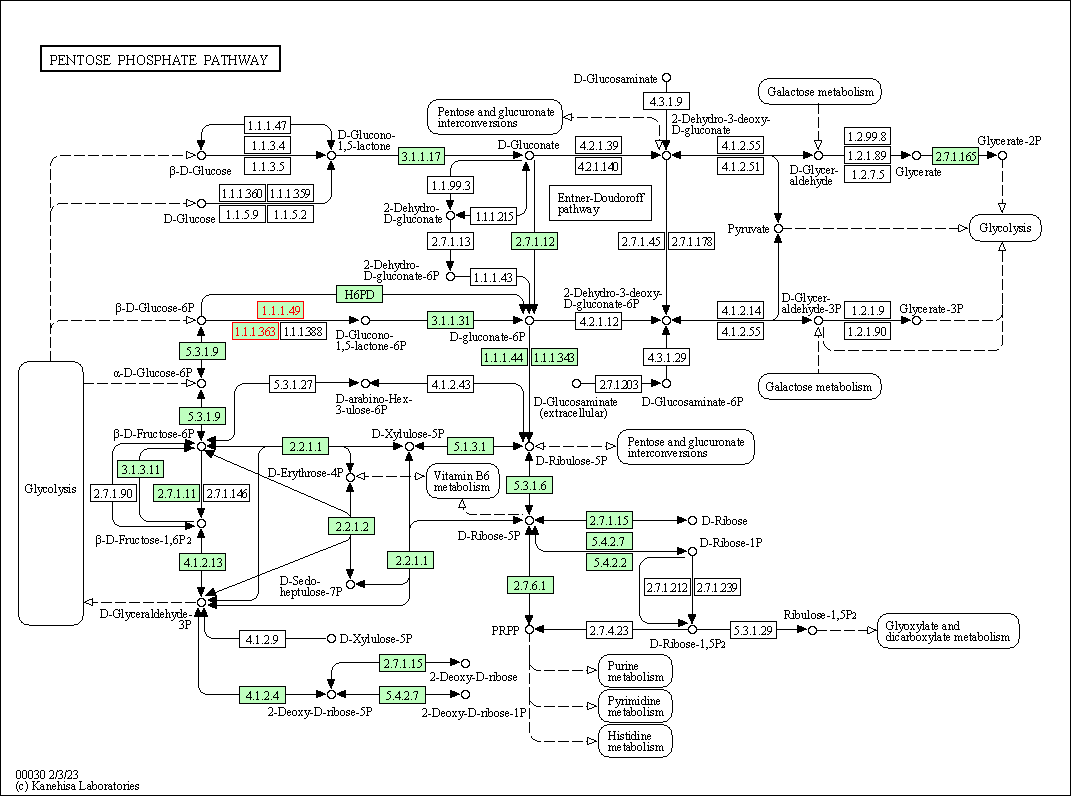
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Glutathione metabolism | hsa00480 | Affiliated Target |
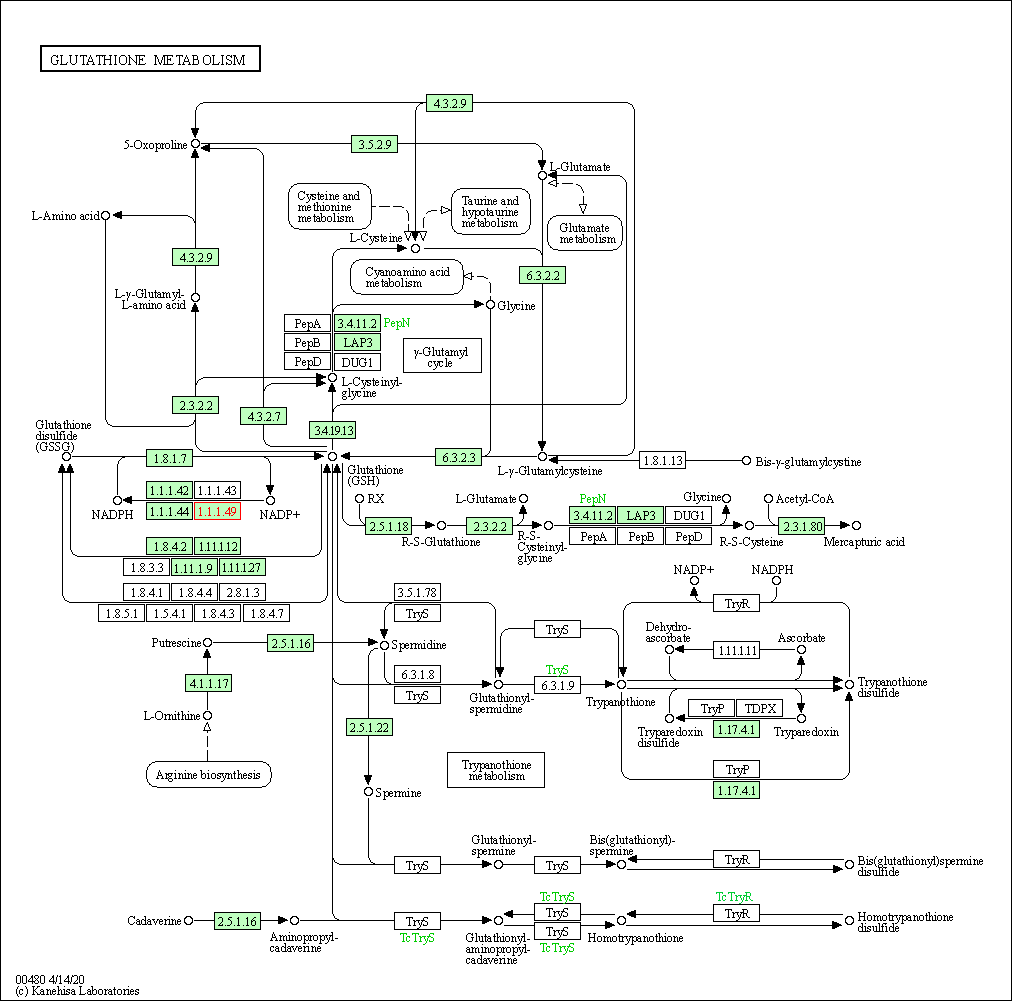
|
| Class: Metabolism => Metabolism of other amino acids | Pathway Hierarchy | ||
| Degree | 11 | Degree centrality | 1.18E-03 | Betweenness centrality | 2.21E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.36E-01 | Radiality | 1.41E+01 | Clustering coefficient | 2.55E-01 |
| Neighborhood connectivity | 4.00E+01 | Topological coefficient | 1.14E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Glucose utilization and activity of glucose-6-phosphate dehydrogenase, isocitrate dehydrogenase and malate dehydrogenase in rat erythrocytes after treatment with tuberculostatic agents. Vopr Med Khim. 1986 Sep-Oct;32(5):32-5. | |||||
| REF 2 | Direct agonist/antagonist functions of dehydroepiandrosterone. Endocrinology. 2005 Nov;146(11):4568-76. | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800007279) | |||||
| REF 4 | Inhibition of Trypanosoma brucei glucose-6-phosphate dehydrogenase by human steroids and their effects on the viability of cultured parasites. Bioorg Med Chem. 2009 Mar 15;17(6):2483-9. | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800007279) | |||||
| REF 6 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 7 | Crystal structure of Human G6PD Canton | |||||
| REF 8 | Allosteric role of a structural NADP(+) molecule in glucose-6-phosphate dehydrogenase activity. Proc Natl Acad Sci U S A. 2022 Jul 19;119(29):e2119695119. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

