Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T65116
(Former ID: TTDR01229)
|
|||||
| Target Name |
Ecto-5'-nucleotidase (CD73)
|
|||||
| Synonyms |
NT5; CD73 antigen; 5'-nucleotidase; 5'-NT
Click to Show/Hide
|
|||||
| Gene Name |
NT5E
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Ovarian cancer [ICD-11: 2C73] | |||||
| 2 | Pancreatic cancer [ICD-11: 2C10] | |||||
| 3 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Exhibits AMP-, NAD-, and NMN-nucleosidase activities. Hydrolyzes extracellular nucleotides into membrane permeable nucleosides.
Click to Show/Hide
|
|||||
| BioChemical Class |
Phosphoric monoester hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.1.3.5
|
|||||
| Sequence |
MCPRAARAPATLLLALGAVLWPAAGAWELTILHTNDVHSRLEQTSEDSSKCVNASRCMGG
VARLFTKVQQIRRAEPNVLLLDAGDQYQGTIWFTVYKGAEVAHFMNALRYDAMALGNHEF DNGVEGLIEPLLKEAKFPILSANIKAKGPLASQISGLYLPYKVLPVGDEVVGIVGYTSKE TPFLSNPGTNLVFEDEITALQPEVDKLKTLNVNKIIALGHSGFEMDKLIAQKVRGVDVVV GGHSNTFLYTGNPPSKEVPAGKYPFIVTSDDGRKVPVVQAYAFGKYLGYLKIEFDERGNV ISSHGNPILLNSSIPEDPSIKADINKWRIKLDNYSTQELGKTIVYLDGSSQSCRFRECNM GNLICDAMINNNLRHTDEMFWNHVSMCILNGGGIRSPIDERNNGTITWENLAAVLPFGGT FDLVQLKGSTLKKAFEHSVHRYGQSTGEFLQVGGIHVVYDLSRKPGDRVVKLDVLCTKCR VPSYDPLKMDEVYKVILPNFLANGGDGFQMIKDELLRHDSGDQDINVVSTYISKMKVIYP AVEGRIKFSTGSHCHGSFSLIFLSLWAVIFVLYQ Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T56TFE | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Oleclumab | Drug Info | Phase 1 | Solid tumour/cancer | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 34 Inhibitor drugs | + | ||||
| 1 | Oleclumab | Drug Info | [1] | |||
| 2 | PMID28870136-Compound-36 | Drug Info | [8] | |||
| 3 | PMID28870136-Compound-38 | Drug Info | [8] | |||
| 4 | PMID28870136-Compound-39 | Drug Info | [8] | |||
| 5 | PMID28870136-Compound-40 | Drug Info | [8] | |||
| 6 | PMID28870136-Compound-41 | Drug Info | [8] | |||
| 7 | PMID28870136-Compound-42 | Drug Info | [8] | |||
| 8 | PMID28870136-Compound-43 | Drug Info | [8] | |||
| 9 | PMID28870136-Compound-44 | Drug Info | [8] | |||
| 10 | PMID28870136-Compound-45 | Drug Info | [8] | |||
| 11 | PMID28870136-Compound-46 | Drug Info | [8] | |||
| 12 | PMID28870136-Compound-47 | Drug Info | [8] | |||
| 13 | PMID28870136-Compound-48 | Drug Info | [8] | |||
| 14 | PMID28870136-Compound-49 | Drug Info | [8] | |||
| 15 | PMID28870136-Compound-50 | Drug Info | [8] | |||
| 16 | PMID28870136-Compound-51 | Drug Info | [8] | |||
| 17 | PMID28870136-Compound-52 | Drug Info | [8] | |||
| 18 | PMID28870136-Compound-53 | Drug Info | [8] | |||
| 19 | PMID28870136-Compound-55 | Drug Info | [8] | |||
| 20 | PMID28870136-Compound-57 | Drug Info | [8] | |||
| 21 | PMID28870136-Compound-59 | Drug Info | [8] | |||
| 22 | PMID28870136-Compound-60 | Drug Info | [8] | |||
| 23 | PMID28870136-Compound-61 | Drug Info | [8] | |||
| 24 | PMID28870136-Compound-64 | Drug Info | [8] | |||
| 25 | PMID28870136-Compound-65 | Drug Info | [8] | |||
| 26 | PMID28870136-Compound-67 | Drug Info | [8] | |||
| 27 | PMID28870136-Compound-68 | Drug Info | [8] | |||
| 28 | PMID28870136-Compound-69 | Drug Info | [8] | |||
| 29 | PMID29166791-Compound-AMPCP | Drug Info | [9] | |||
| 30 | Acid blue 25 | Drug Info | [10] | |||
| 31 | alphabeta-methyleneADP | Drug Info | [11] | |||
| 32 | PSB-0952 | Drug Info | [10] | |||
| 33 | PSB-0963 | Drug Info | [10] | |||
| 34 | RB 2 | Drug Info | [10] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Riboflavin | Ligand Info | |||||
| Structure Description | Crystal structure of CD73 in complex with riboflavin in the open form | PDB:7PD9 | ||||
| Method | X-ray diffraction | Resolution | 1.39 Å | Mutation | Yes | [12] |
| PDB Sequence |
PWELTILHTN
35 DVHSRLEQTS45 EDSSKCVDAS55 RCMGGVARLF65 TKVQQIRRAE75 PNVLLLDAGD 85 QYQGTIWFTV95 YKGAEVAHFM105 NALRYDAMAL115 GNHEFDNGVE125 GLIEPLLKEA 135 KFPILSANIS145 ASGPLASQIS155 GLYLPYKVLP165 VGDEVVGIVG175 YTSKETPFLS 185 NPGTNLVFED195 EITALQPEVD205 KLKTLNVNKI215 IALGHSGFEM225 DKLIAQKVRG 235 VDVVVGGHSN245 TFLYTGNPPS255 KEVPAGKYPF265 IVTSDDGRKV275 PVVQAYAFGK 285 YLGYLKIEFD295 ERGNVISSHG305 NPILLDSSIP315 EDPSIKADIN325 KWRIKLDDYS 335 TQELGKTIVY345 LDGSSQSCRF355 RECNMGNLIC365 DAMINNNLRH375 ADEMFWNHVS 385 MCILNGGGIR395 SPIDERNDGT405 ITWENLAAVL415 PFGGTFDLVQ425 LKGSTLKKAF 435 EHSVHRYGQS445 TGEFLQVGGI455 HVVYDLSRKP465 GDRVVKLDVL475 CTSCRVPSYD 485 PLKMDEVYKV495 ILPNFLANGG505 DGFQMIKDEL515 LRHDSGDQDI525 NVVSTYISKM 535 KVIYPAVEGR545 IKFS
|
|||||
|
|
||||||
| Ligand Name: Caffeine | Ligand Info | |||||
| Structure Description | Crystal structure of CD73 in complex with caffeine in the open form | PDB:7PBB | ||||
| Method | X-ray diffraction | Resolution | 1.47 Å | Mutation | Yes | [12] |
| PDB Sequence |
PWELTILHTN
35 DVHSRLEQTS45 EDSSKCVDAS55 RCMGGVARLF65 TKVQQIRRAE75 PNVLLLDAGD 85 QYQGTIWFTV95 YKGAEVAHFM105 NALRYDAMAL115 GNHEFDNGVE125 GLIEPLLKEA 135 KFPILSANIS145 ASGPLASQIS155 GLYLPYKVLP165 VGDEVVGIVG175 YTSKETPFLS 185 NPGTNLVFED195 EITALQPEVD205 KLKTLNVNKI215 IALGHSGFEM225 DKLIAQKVRG 235 VDVVVGGHSN245 TFLYTGNPPS255 KEVPAGKYPF265 IVTSDDGRKV275 PVVQAYAFGK 285 YLGYLKIEFD295 ERGNVISSHG305 NPILLDSSIP315 EDPSIKADIN325 KWRIKLDDYS 335 TQELGKTIVY345 LDGSSQSCRF355 RECNMGNLIC365 DAMINNNLRH375 ADEMFWNHVS 385 MCILNGGGIR395 SPIDERNDGT405 ITWENLAAVL415 PFGGTFDLVQ425 LKGSTLKKAF 435 EHSVHRYGQS445 TGEFLQVGGI455 HVVYDLSRKP465 GDRVVKLDVL475 CTSCRVPSYD 485 PLKMDEVYKV495 ILPNFLANGG505 DGFQMIKDEL515 LRHDSGDQDI525 NVVSTYISKM 535 KVIYPAVEGR545 IKFS
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Purine metabolism | hsa00230 | Affiliated Target |
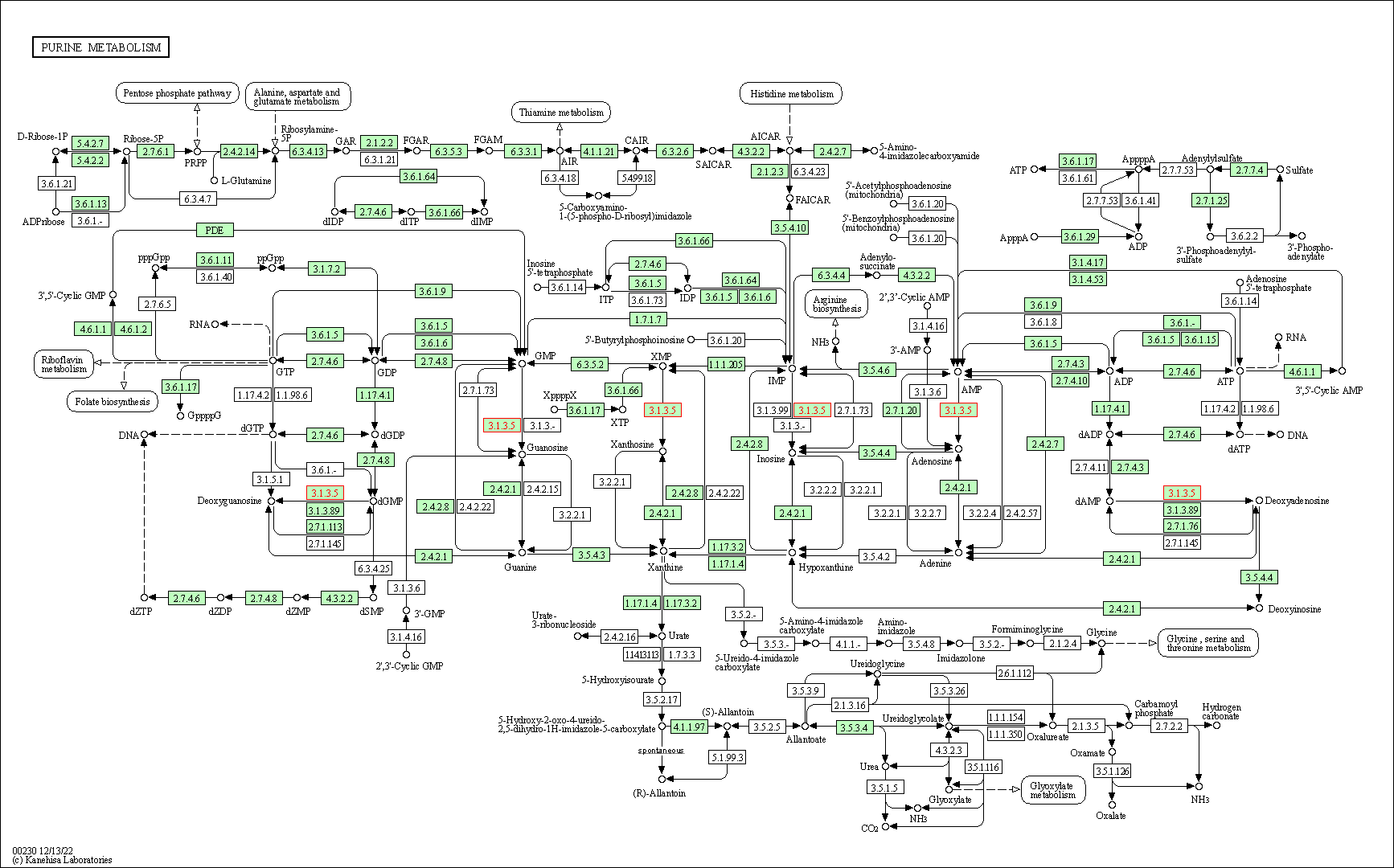
|
| Class: Metabolism => Nucleotide metabolism | Pathway Hierarchy | ||
| Pyrimidine metabolism | hsa00240 | Affiliated Target |
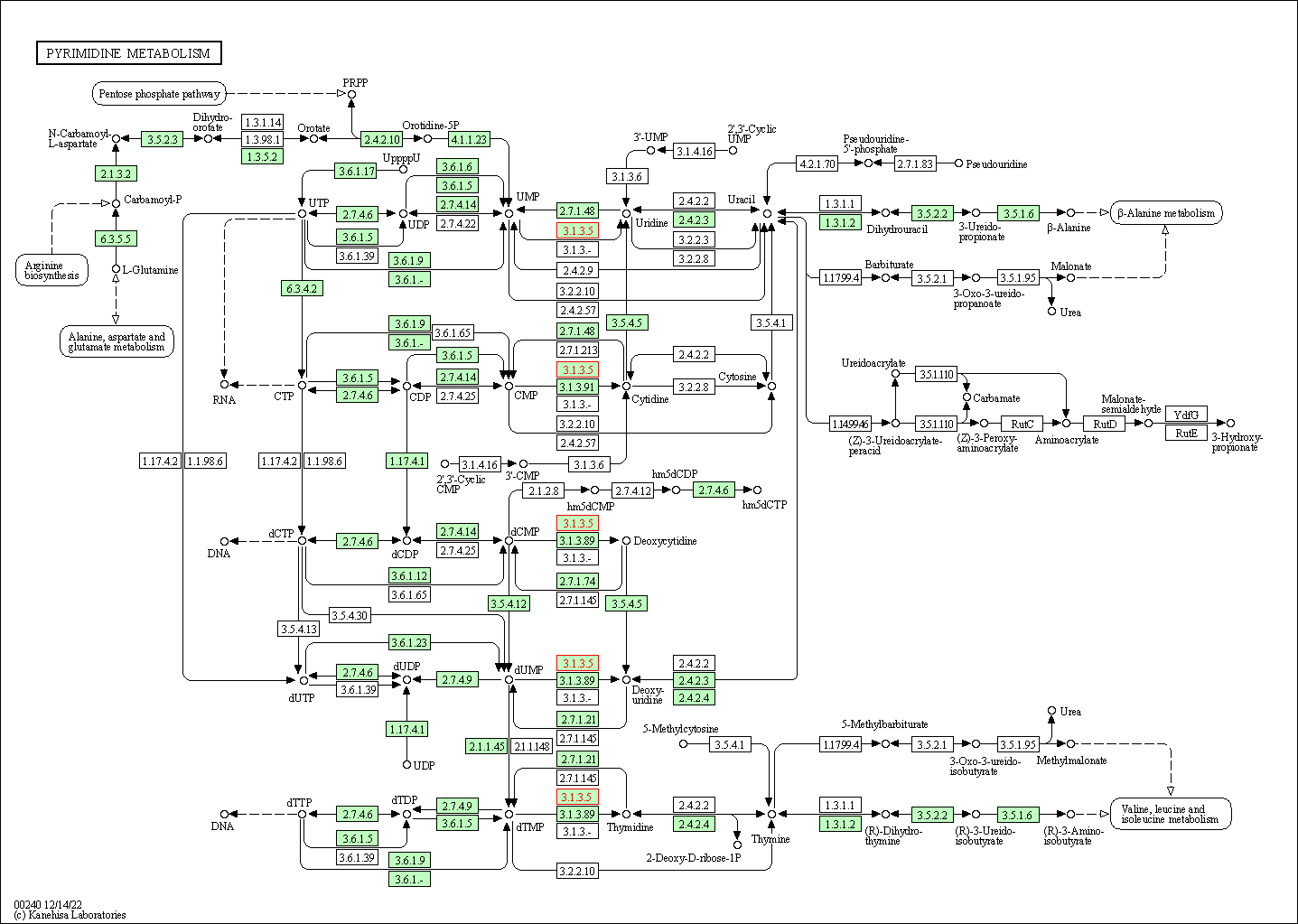
|
| Class: Metabolism => Nucleotide metabolism | Pathway Hierarchy | ||
| Nicotinate and nicotinamide metabolism | hsa00760 | Affiliated Target |
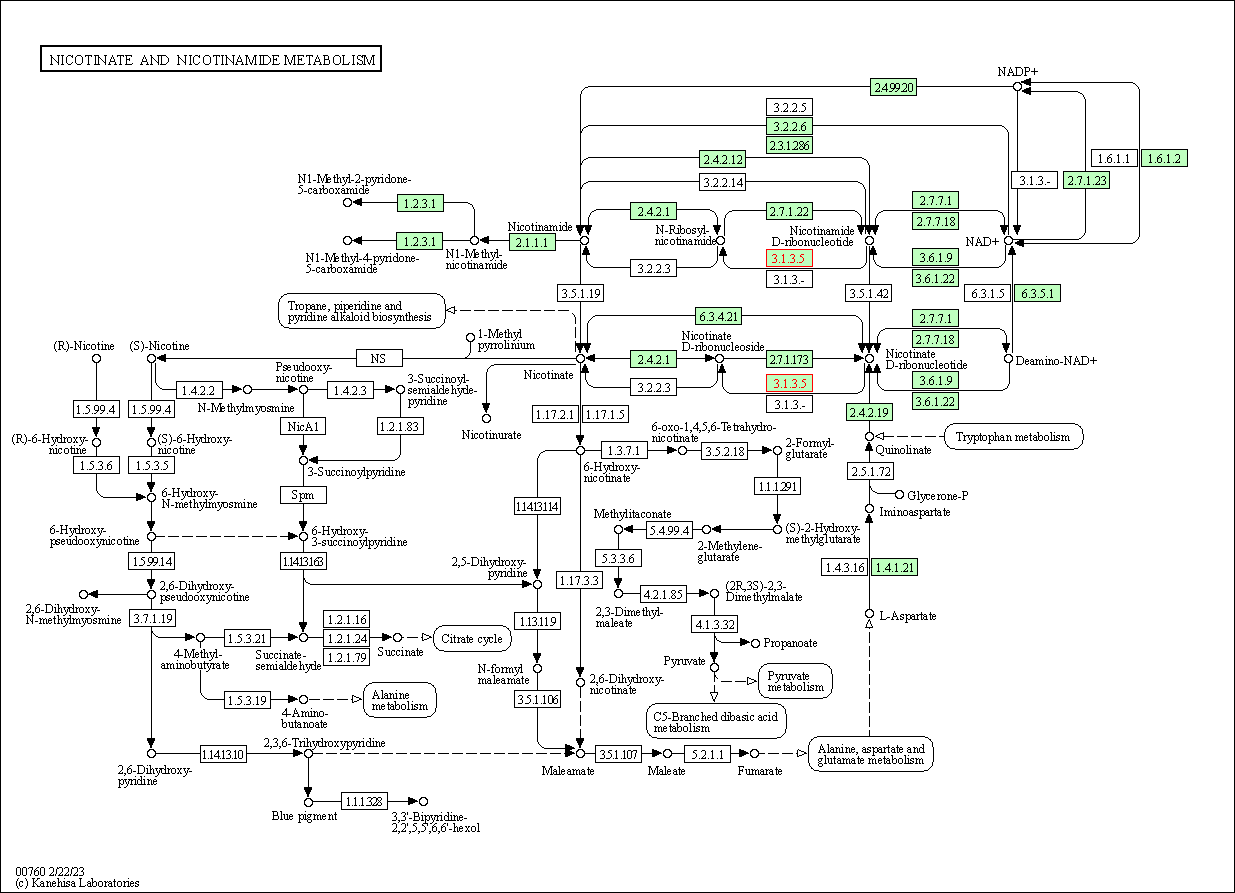
|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Degree | 3 | Degree centrality | 3.22E-04 | Betweenness centrality | 4.55E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.59E-01 | Radiality | 1.23E+01 | Clustering coefficient | 6.67E-01 |
| Neighborhood connectivity | 8.33E+00 | Topological coefficient | 4.17E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 3 BioCyc Pathways | + | ||||
| 1 | Purine nucleotides degradation | |||||
| 2 | Urate biosynthesis/inosine 5'-phosphate degradation | |||||
| 3 | Adenosine nucleotides degradation | |||||
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Purine metabolism | |||||
| 2 | Pyrimidine metabolism | |||||
| 3 | Nicotinate and nicotinamide metabolism | |||||
| 4 | Metabolic pathways | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Purine metabolism | |||||
| 2 | Pyrimidine Metabolism | |||||
| PID Pathway | [+] 1 PID Pathways | + | ||||
| 1 | HIF-1-alpha transcription factor network | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Purine catabolism | |||||
| WikiPathways | [+] 5 WikiPathways | + | ||||
| 1 | Differentiation Pathway | |||||
| 2 | miR-targeted genes in muscle cell - TarBase | |||||
| 3 | miR-targeted genes in lymphocytes - TarBase | |||||
| 4 | miR-targeted genes in epithelium - TarBase | |||||
| 5 | Metabolism of nucleotides | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | ClinicalTrials.gov (NCT05001347) A Phase 2 Clinical Study of TJ004309 in Combination With Atezolizumab (TECENTRIQ?) in Patients With Advanced or Metastatic Ovarian Cancers and Selected Advanced Solid Tumors. U.S.National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT04322006) A Phase I/II Study of TJ004309 for Advanced Solid Tumor. U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT03549000) A Phase I/Ib Study of NZV930 Alone and in Combination With PDR001 and /or NIR178 in Patients With Advanced Malignancies.. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT04148937) A Study of the CD73 Inhibitor LY3475070 Alone or in Combination With Pembrolizumab in Participants With Advanced Cancer. U.S. National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT04104672) A Study to Evaluate the Safety and Tolerability of AB680 in Participants With Gastrointestinal Malignancies. U.S. National Institutes of Health. | |||||
| REF 7 | ClinicalTrials.gov (NCT03954704) Study of GS-1423 in Participants With Advanced Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 8 | Ectonucleotidase inhibitors: a patent review (2011-2016).Expert Opin Ther Pat. 2017 Dec;27(12):1291-1304. | |||||
| REF 9 | Evaluation of WO2017098421: GSK's benzothiazine compounds as CD73 inhibitor filings.Expert Opin Ther Pat. 2018 Feb;28(2):167-171. | |||||
| REF 10 | Development of potent and selective inhibitors of ecto-5'-nucleotidase based on an anthraquinone scaffold. J Med Chem. 2010 Mar 11;53(5):2076-86. | |||||
| REF 11 | 5'-Nucleotidase from smooth muscle of small intestine and from brain. Inhibition of nucleotides. Biochemistry. 1975 Jun 3;14(11):2362-6. | |||||
| REF 12 | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives. Purinergic Signal. 2021 Dec;17(4):693-704. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

