Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T19579
(Former ID: TTDC00026)
|
|||||
| Target Name |
Interleukin 2 receptor beta (IL2RB)
|
|||||
| Synonyms |
P70-75; Interleukin-2 receptor subunit beta; Interleukin-15 receptor subunit beta; IL15RB; IL-2RB; IL-2R subunit beta; IL-2 receptor subunit beta; IL-2 receptor; High affinity IL-2 receptor subunit beta; CD122
Click to Show/Hide
|
|||||
| Gene Name |
IL2RB
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Melanoma [ICD-11: 2C30] | |||||
| Function |
Receptor for interleukin-2. This beta subunit is involved in receptor mediated endocytosis and transduces the mitogenic signals of IL2. Probably in association with IL15RA, involved in the stimulation of neutrophil phagocytosis by IL15.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine receptor
|
|||||
| UniProt ID | ||||||
| Sequence |
MAAPALSWRLPLLILLLPLATSWASAAVNGTSQFTCFYNSRANISCVWSQDGALQDTSCQ
VHAWPDRRRWNQTCELLPVSQASWACNLILGAPDSQKLTTVDIVTLRVLCREGVRWRVMA IQDFKPFENLRLMAPISLQVVHVETHRCNISWEISQASHYFERHLEFEARTLSPGHTWEE APLLTLKQKQEWICLETLTPDTQYEFQVRVKPLQGEFTTWSPWSQPLAFRTKPAALGKDT IPWLGHLLVGLSGAFGFIILVYLLINCRNTGPWLKKVLKCNTPDPSKFFSQLSSEHGGDV QKWLSSPFPSSSFSPGGLAPEISPLEVLERDKVTQLLLQQDKVPEPASLSSNHSLTSCFT NQGYFFFHLPDALEIEACQVYFTYDPYSEEDPDEGVAGAPTGSSPQPLQPLSGEDDAYCT FPSRDDLLLFSPSLLGGPSPPSTAPGGSGAGEERMPPSLQERVPRDWDPQPLGPPTPGVP DLVDFQPPPELVLREAGEEVPDAGPREGVSFPWSRPPGQGEFRALNARLPLNTDAYLSLQ ELQGQDPTHLV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | NKTR 214 | Drug Info | Phase 3 | Melanoma | [3] | |
| 2 | AFTVac | Drug Info | Phase 2 | Glioblastoma multiforme | [1] | |
| 3 | Leukocyte interleukin | Drug Info | Phase 1 | Human immunodeficiency virus infection | [4] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Agonist | [+] 1 Agonist drugs | + | ||||
| 1 | NKTR 214 | Drug Info | [5] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | Leukocyte interleukin | Drug Info | [6] | |||
| 2 | DAB-486-IL-2 | Drug Info | [7] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Alpha-D-Mannose | Ligand Info | |||||
| Structure Description | Crystal structure of a interleukin-2 variant in complex with interleukin-2 receptor | PDB:5M5E | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | No | [8] |
| PDB Sequence |
SQFTCFYNSR
15 ANISCVWSQD25 GALQDTSCQV35 HAWPDRRRWN45 QTCELLPVSQ55 ASWACNLILG 65 APDSQKLTTV75 DIVTLRVLCR85 EGVRWRVMAI95 QDFKPFENLR105 LMAPISLQVV 115 HVETHRCNIS125 WEISQASHYF135 ERHLEFEART145 LSPGHTWEEA155 PLLTLKQKQE 165 WICLETLTPD175 TQYEFQVRVK185 PLQGEFTTWS195 PWSQPLAFRT205 KPA |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Interleukin 21 receptor (IL21R) | 26.582 (84/316) | 7.99E-13 | |
| Interleukin-9 receptor (IL9R) | 26.481 (76/287) | 7.19E-11 | |
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |
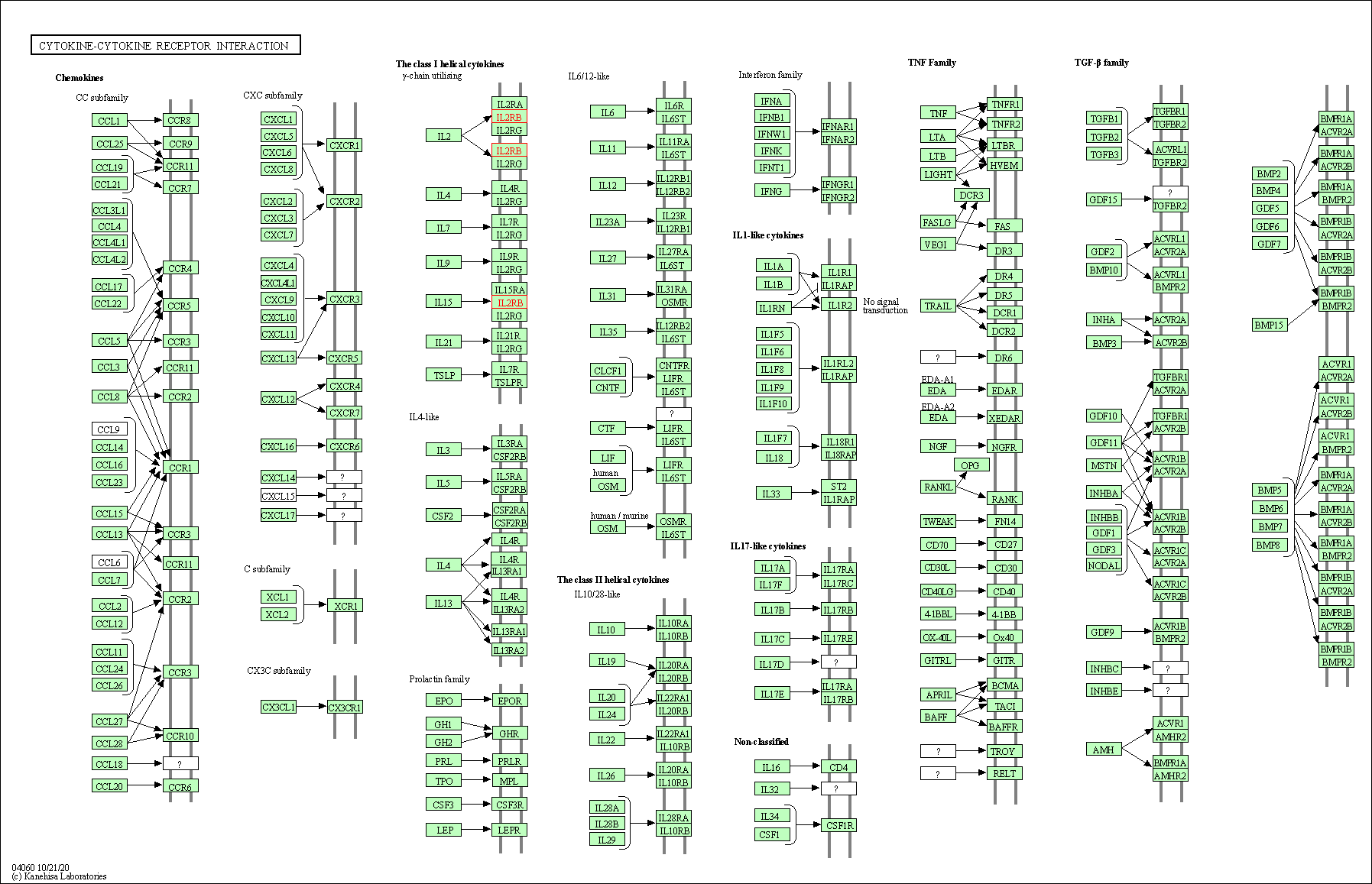
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Viral protein interaction with cytokine and cytokine receptor | hsa04061 | Affiliated Target |
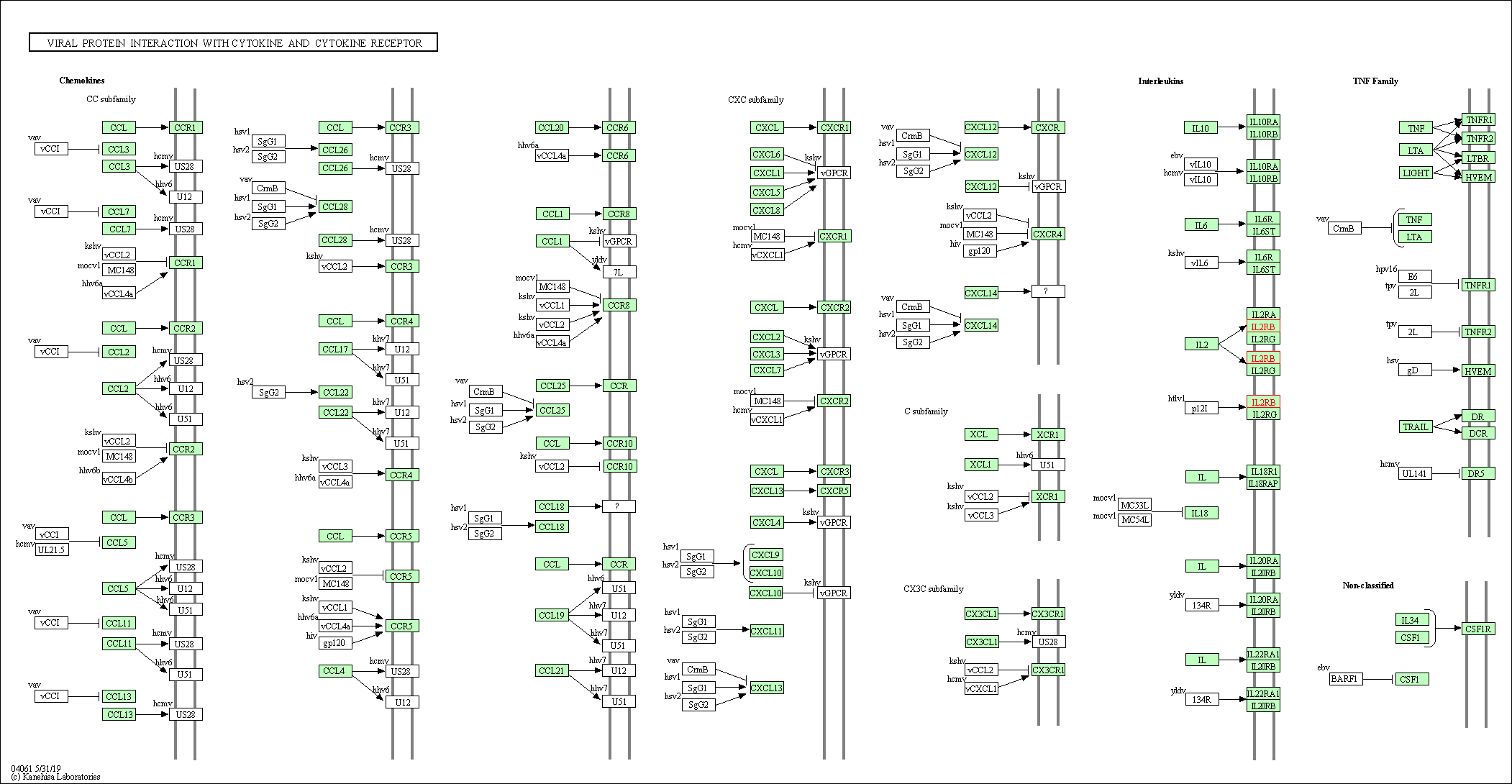
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |
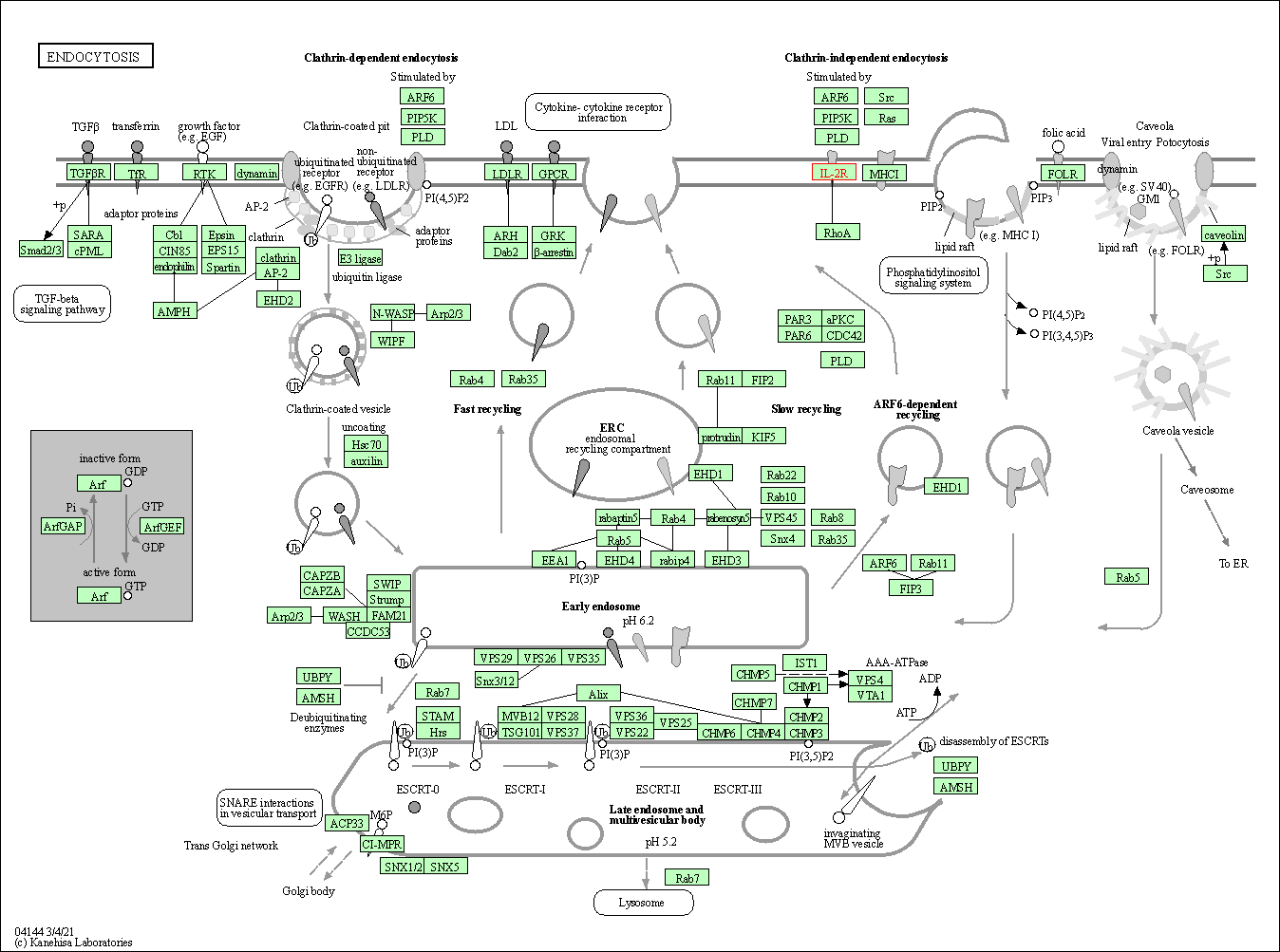
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
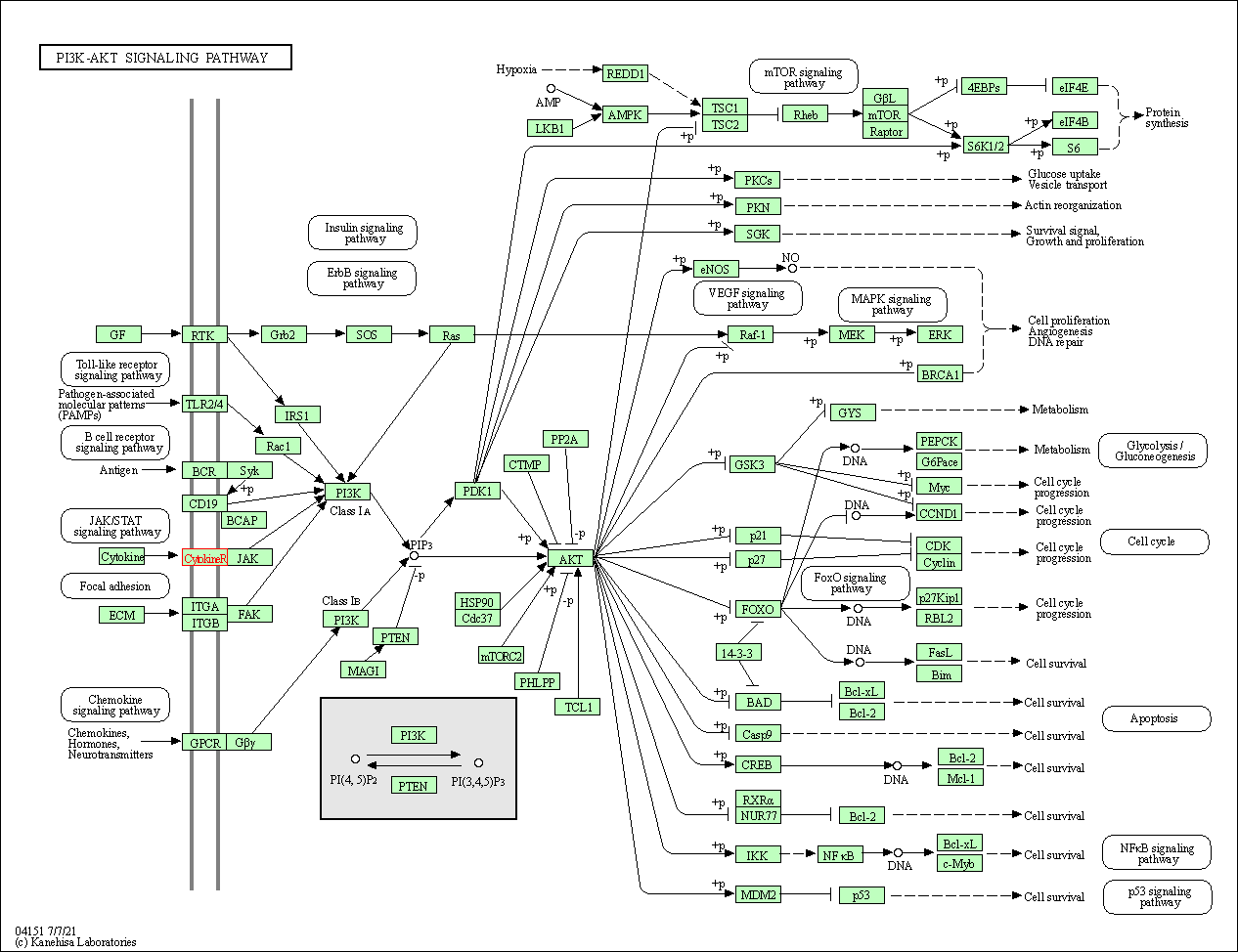
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| JAK-STAT signaling pathway | hsa04630 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Th1 and Th2 cell differentiation | hsa04658 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 15 | Degree centrality | 1.61E-03 | Betweenness centrality | 1.28E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.28E-01 | Radiality | 1.40E+01 | Clustering coefficient | 4.86E-01 |
| Neighborhood connectivity | 4.09E+01 | Topological coefficient | 1.45E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating Transcription Factors | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 7 KEGG Pathways | + | ||||
| 1 | Cytokine-cytokine receptor interaction | |||||
| 2 | Endocytosis | |||||
| 3 | PI3K-Akt signaling pathway | |||||
| 4 | Jak-STAT signaling pathway | |||||
| 5 | Measles | |||||
| 6 | HTLV-I infection | |||||
| 7 | Transcriptional misregulation in cancer | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | IL2 Signaling Pathway | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Interleukin signaling pathway | |||||
| PID Pathway | [+] 7 PID Pathways | + | ||||
| 1 | IL12-mediated signaling events | |||||
| 2 | SHP2 signaling | |||||
| 3 | IL2-mediated signaling events | |||||
| 4 | IL2 signaling events mediated by PI3K | |||||
| 5 | IL2 signaling events mediated by STAT5 | |||||
| 6 | Downstream signaling in naï | |||||
| 7 | ||||||
| Reactome | [+] 5 Reactome Pathways | + | ||||
| 1 | GPVI-mediated activation cascade | |||||
| 2 | G beta:gamma signalling through PI3Kgamma | |||||
| 3 | Interleukin-2 signaling | |||||
| 4 | RAF/MAP kinase cascade | |||||
| 5 | Interleukin receptor SHC signaling | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | IL-2 Signaling Pathway | |||||
| 2 | Inflammatory Response Pathway | |||||
| 3 | Interleukin-2 signaling | |||||
| 4 | Interleukin-3, 5 and GM-CSF signaling | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Phase II randomized trial of autologous formalin-fixed tumor vaccine for postsurgical recurrence of hepatocellular carcinoma. Clin Cancer Res. 2004 Mar 1;10(5):1574-9. | |||||
| REF 2 | ClinicalTrials.gov (NCT03635983) A Study of NKTR-214 Combined With Nivolumab vs Nivolumab Alone in Participants With Previously Untreated Inoperable or Metastatic Melanoma. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT03635983) A Study of NKTR-214 Combined With Nivolumab vs Nivolumab Alone in Participants With Previously Untreated Inoperable or Metastatic Melanoma. U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT02115919) Safety Study of Multikine in the Treatment of Perianal Warts. U.S. National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 6 | Leukocyte Interleukin, Inj. (LI) augmentation of natural killer cells and cytolytic T-lymphocytes. Immunopharmacol Immunotoxicol. 1995 May;17(2):247-64. | |||||
| REF 7 | Interleukin 2 receptor targeted fusion toxin (DAB486-IL-2) treatment blocks diabetogenic autoimmunity in non-obese diabetic mice. Eur J Immunol. 1992 Mar;22(3):697-702. | |||||
| REF 8 | Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines. Oncoimmunology. 2017 Jan 11;6(3):e1277306. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

