Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T97196
(Former ID: TTDI03045)
|
|||||
| Target Name |
ATM serine/threonine kinase (ATM)
|
|||||
| Synonyms |
Serine-protein kinase ATM; Ataxia telangiectasia mutated; A-T mutated
Click to Show/Hide
|
|||||
| Gene Name |
ATM
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Brain cancer [ICD-11: 2A00] | |||||
| 2 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| 3 | Lung cancer [ICD-11: 2C25] | |||||
| Function |
Serine/threonine protein kinase which activates checkpoint signaling upon double strand breaks (DSBs), apoptosis and genotoxic stresses such as ionizing ultraviolet A light (UVA), thereby acting as a DNA damage sensor. Recognizes the substrate consensus sequence [ST]-Q. Phosphorylates 'Ser-139' of histone variant H2AX/H2AFX at double strand breaks (DSBs), thereby regulating DNA damage response mechanism. Also plays a role in pre-B cell allelic exclusion, a process leading to expression of a single immunoglobulin heavy chain allele to enforce clonality and monospecific recognition by the B-cell antigen receptor (BCR) expressed on individual B-lymphocytes. After the introduction of DNA breaks by the RAG complex on one immunoglobulin allele, acts by mediating a repositioning of the second allele to pericentromeric heterochromatin, preventing accessibility to the RAG complex and recombination of the second allele. Also involved in signal transduction and cell cycle control. May function as a tumor suppressor. Necessary for activation of ABL1 and SAPK. Phosphorylates DYRK2, CHEK2, p53/TP53, FANCD2, NFKBIA, BRCA1, CTIP, nibrin (NBN), TERF1, RAD9, UBQLN4 and DCLRE1C. May play a role in vesicle and/or protein transport. Could play a role in T-cell development, gonad and neurological function. Plays a role in replication-dependent histone mRNA degradation. Binds DNA ends. Phosphorylation of DYRK2 in nucleus in response to genotoxic stress prevents its MDM2-mediated ubiquitination and subsequent proteasome degradation. Phosphorylates ATF2 which stimulates its function in DNA damage response.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MSLVLNDLLICCRQLEHDRATERKKEVEKFKRLIRDPETIKHLDRHSDSKQGKYLNWDAV
FRFLQKYIQKETECLRIAKPNVSASTQASRQKKMQEISSLVKYFIKCANRRAPRLKCQEL LNYIMDTVKDSSNGAIYGADCSNILLKDILSVRKYWCEISQQQWLELFSVYFRLYLKPSQ DVHRVLVARIIHAVTKGCCSQTDGLNSKFLDFFSKAIQCARQEKSSSGLNHILAALTIFL KTLAVNFRIRVCELGDEILPTLLYIWTQHRLNDSLKEVIIELFQLQIYIHHPKGAKTQEK GAYESTKWRSILYNLYDLLVNEISHIGSRGKYSSGFRNIAVKENLIELMADICHQVFNED TRSLEISQSYTTTQRESSDYSVPCKRKKIELGWEVIKDHLQKSQNDFDLVPWLQIATQLI SKYPASLPNCELSPLLMILSQLLPQQRHGERTPYVLRCLTEVALCQDKRSNLESSQKSDL LKLWNKIWCITFRGISSEQIQAENFGLLGAIIQGSLVEVDREFWKLFTGSACRPSCPAVC CLTLALTTSIVPGTVKMGIEQNMCEVNRSFSLKESIMKWLLFYQLEGDLENSTEVPPILH SNFPHLVLEKILVSLTMKNCKAAMNFFQSVPECEHHQKDKEELSFSEVEELFLQTTFDKM DFLTIVRECGIEKHQSSIGFSVHQNLKESLDRCLLGLSEQLLNNYSSEITNSETLVRCSR LLVGVLGCYCYMGVIAEEEAYKSELFQKAKSLMQCAGESITLFKNKTNEEFRIGSLRNMM QLCTRCLSNCTKKSPNKIASGFFLRLLTSKLMNDIADICKSLASFIKKPFDRGEVESMED DTNGNLMEVEDQSSMNLFNDYPDSSVSDANEPGESQSTIGAINPLAEEYLSKQDLLFLDM LKFLCLCVTTAQTNTVSFRAADIRRKLLMLIDSSTLEPTKSLHLHMYLMLLKELPGEEYP LPMEDVLELLKPLSNVCSLYRRDQDVCKTILNHVLHVVKNLGQSNMDSENTRDAQGQFLT VIGAFWHLTKERKYIFSVRMALVNCLKTLLEADPYSKWAILNVMGKDFPVNEVFTQFLAD NHHQVRMLAAESINRLFQDTKGDSSRLLKALPLKLQQTAFENAYLKAQEGMREMSHSAEN PETLDEIYNRKSVLLTLIAVVLSCSPICEKQALFALCKSVKENGLEPHLVKKVLEKVSET FGYRRLEDFMASHLDYLVLEWLNLQDTEYNLSSFPFILLNYTNIEDFYRSCYKVLIPHLV IRSHFDEVKSIANQIQEDWKSLLTDCFPKILVNILPYFAYEGTRDSGMAQQRETATKVYD MLKSENLLGKQIDHLFISNLPEIVVELLMTLHEPANSSASQSTDLCDFSGDLDPAPNPPH FPSHVIKATFAYISNCHKTKLKSILEILSKSPDSYQKILLAICEQAAETNNVYKKHRILK IYHLFVSLLLKDIKSGLGGAWAFVLRDVIYTLIHYINQRPSCIMDVSLRSFSLCCDLLSQ VCQTAVTYCKDALENHLHVIVGTLIPLVYEQVEVQKQVLDLLKYLVIDNKDNENLYITIK LLDPFPDHVVFKDLRITQQKIKYSRGPFSLLEEINHFLSVSVYDALPLTRLEGLKDLRRQ LELHKDQMVDIMRASQDNPQDGIMVKLVVNLLQLSKMAINHTGEKEVLEAVGSCLGEVGP IDFSTIAIQHSKDASYTKALKLFEDKELQWTFIMLTYLNNTLVEDCVKVRSAAVTCLKNI LATKTGHSFWEIYKMTTDPMLAYLQPFRTSRKKFLEVPRFDKENPFEGLDDINLWIPLSE NHDIWIKTLTCAFLDSGGTKCEILQLLKPMCEVKTDFCQTVLPYLIHDILLQDTNESWRN LLSTHVQGFFTSCLRHFSQTSRSTTPANLDSESEHFFRCCLDKKSQRTMLAVVDYMRRQK RPSSGTIFNDAFWLDLNYLEVAKVAQSCAAHFTALLYAEIYADKKSMDDQEKRSLAFEEG SQSTTISSLSEKSKEETGISLQDLLLEIYRSIGEPDSLYGCGGGKMLQPITRLRTYEHEA MWGKALVTYDLETAIPSSTRQAGIIQALQNLGLCHILSVYLKGLDYENKDWCPELEELHY QAAWRNMQWDHCTSVSKEVEGTSYHESLYNALQSLRDREFSTFYESLKYARVKEVEEMCK RSLESVYSLYPTLSRLQAIGELESIGELFSRSVTHRQLSEVYIKWQKHSQLLKDSDFSFQ EPIMALRTVILEILMEKEMDNSQRECIKDILTKHLVELSILARTFKNTQLPERAIFQIKQ YNSVSCGVSEWQLEEAQVFWAKKEQSLALSILKQMIKKLDASCAANNPSLKLTYTECLRV CGNWLAETCLENPAVIMQTYLEKAVEVAGNYDGESSDELRNGKMKAFLSLARFSDTQYQR IENYMKSSEFENKQALLKRAKEEVGLLREHKIQTNRYTVKVQRELELDELALRALKEDRK RFLCKAVENYINCLLSGEEHDMWVFRLCSLWLENSGVSEVNGMMKRDGMKIPTYKFLPLM YQLAARMGTKMMGGLGFHEVLNNLISRISMDHPHHTLFIILALANANRDEFLTKPEVARR SRITKNVPKQSSQLDEDRTEAANRIICTIRSRRPQMVRSVEALCDAYIILANLDATQWKT QRKGINIPADQPITKLKNLEDVVVPTMEIKVDHTGEYGNLVTIQSFKAEFRLAGGVNLPK IIDCVGSDGKERRQLVKGRDDLRQDAVMQQVFQMCNTLLQRNTETRKRKLTICTYKVVPL SQRSGVLEWCTGTVPIGEFLVNNEDGAHKRYRPNDFSAFQCQKKMMEVQKKSFEEKYEVF MDVCQNFQPVFRYFCMEKFLDPAIWFEKRLAYTRSVATSSIVGYILGLGDRHVQNILINE QSAELVHIDLGVAFEQGKILPTPETVPFRLTRDIVDGMGITGVEGVFRRCCEKTMEVMRN SQETLLTIVEVLLYDPLFDWTMNPLKALYLQQRPEDETELHPTLNADDQECKRNLSDIDQ SFNKVAERVLMRLQEKLKGVEEGTVLSVGGQVNLLIQQAIDPKNLSRLFPGWKAWV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T20NCI | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | AZD1390 | Drug Info | Phase 1 | Recurrent glioblastoma | [2] | |
| 2 | M3541 | Drug Info | Phase 1 | Solid tumour/cancer | [2] | |
| 3 | KU-60019 | Drug Info | Clinical trial | Non-small-cell lung cancer | [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 4 Inhibitor drugs | + | ||||
| 1 | AZD1390 | Drug Info | [2] | |||
| 2 | M3541 | Drug Info | [2] | |||
| 3 | KU-60019 | Drug Info | [3] | |||
| 4 | CGK733 | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: AMP-PNP | Ligand Info | |||||
| Structure Description | Human ATM Dimer | PDB:7SIC | ||||
| Method | Electron microscopy | Resolution | 2.51 Å | Mutation | No | [5] |
| PDB Sequence |
LNDLLICCRQ
14 LEHDRATERK24 KEVEKFKRLI34 RDPETIKHLD44 NWDAVFRFLQ65 KYIQKETECQ 87 ASRQKKMQEI97 SSLVKYFIKC107 ANRRAPRLKC117 QELLNYIMDT127 VKDSSNGAIY 137 GADCSNILLK147 DILSVRKYWC157 EISQQQWLEL167 FSVYFRLYLK177 PSQDVHRVLV 187 ARIIHAVTKG197 CCSQTDGLNS207 KFLDFFSKAI217 QCARQEKSSS227 GLNHILAALT 237 IFLKTLAVNF247 RIRVCELGDE257 ILPTLLYIWT267 QHRLNDSLKE277 VIIELFQLQI 287 YIHHPKGAKT297 QEKGAYESTK307 WRSILYNLYD317 LLVNEISHIG327 SRRNIAVKEN 344 LIELMADICH354 QVFNIELGWE394 VIKDHLQKSQ404 NDFDLVPWLQ414 IATQLISKYP 424 ASLPNCELSP434 LLMILSQLLP444 QQRHGERTPY454 VLRCLTEVAL464 CQDKRSNLES 474 SQKSDLLKLW484 NKIWCITFRG494 ISSEQIQAEN504 FGLLGAIIQG514 SLVEVDREFW 524 KLFTGSACRP534 SCPAVCCLTL544 ALTTSIVPGT554 FSLKESIMKW579 LLFYQLETEV 595 PPILHSNFPH605 LVLEKILVSL615 TMKNCKAAMN625 FFQSVPECSF645 SEVEELFLQT 655 TFDKMDFLTG679 FSVHQNLKES689 LDRCLLGLSE699 QLLNNYSSEI709 TNSETLVRCS 719 RLLVGVLGCY729 CYMGVIAEEE739 AYKSELFQKA749 KSLMQCAGES759 ITLFKNKTNE 769 EFRIGSLRNM779 MQLCTRCLSN789 CTKKSPNKIA799 SGFFLRLLTS809 KLMNDIADIC 819 KSLASFISTI879 GAINPLAEEY889 LSKQDLLFLD899 MLKFLCLCVT909 TAQTNTVSFR 919 AADIRRKLLM929 LIDSSTLEPT939 KSLHLHMYLM949 LLKELPGEEY959 PLPMEDVLEL 969 LKPLSNVCSL979 YRRDQDVCKT989 ILNHVLHVVK999 NLGQSNMDSE1009 NTRDAQGQFL 1019 TVIGAFWHLT1029 KERKYIFSVR1039 MALVNCLKTL1049 LEADPYSKWA1059 ILNVMGKDFP 1069 VNEVFTQFLA1079 DNHHQVRMLA1089 AESINRLFQD1099 TKLKALPLKL1115 QQTAFENAYL 1125 KAQEGMREME1142 TLDEIYNRKS1152 VLLTLIAVVL1162 SCSPICEKQA1172 LFALCKSVKE 1182 NGLEPHLVKK1192 VLEKVSETFG1202 YRRLEDFMAS1212 HLDYLVLEWL1222 NLQDTEYNLS 1232 SFPFILLNYT1242 NIEDFYRSCY1252 KVLIPHLVIR1262 SHFDEVKSIA1272 NQIQEDWKSL 1282 LTDCFPKILV1292 NILPYFAYEG1302 TRDSGMAQQR1312 ETATKVYDML1322 KSENLLGKQI 1332 DHLFISNLPE1342 IVVELLMTLH1352 EPDLDPAPNP1378 PHFPSHVIKA1388 TFAYISNCHK 1398 TKLKSILEIL1408 SKSPDSYQKI1418 LLAICEQAAE1428 TNNVYKKHRI1438 LKIYHLFVSL 1448 LLKDIKSGLG1458 GAWAFVLRDV1468 IYTLIHYINQ1478 RPSCIMDVSL1488 RSFSLCCDLL 1498 SQVCQTAVTY1508 CKDALENHLH1518 VIVGTLIPLV1528 YEQVEVQKQV1538 LDLLKYLVID 1548 NKDNENLYIT1558 IKLLDPFPDH1568 VVFKDLRITQ1578 QKIKYSRGPF1588 SLLEEINHFL 1598 SVSVYDALPL1608 TRLEGLKDLR1618 RQLELHKDQM1628 VDIMRASQDN1638 PQDGIMVKLV 1648 VNLLQLSKMA1658 INHTGEKEVL1668 EAVGSCLGEV1678 GPIDFSTIAI1688 QHSKDASYTK 1698 ALKLFEDKEL1708 QWTFIMLTYL1718 NNTLVEDCVK1728 VRSAAVTCLK1738 NILATKTGHS 1748 FWEIYKMTTD1758 PMLAYLQPFR1768 TSRKKFLEVP1778 RFDKENPFEG1788 LDDINLWIPL 1798 SENHDIWIKT1808 LTCAFLDSGG1818 TKCEILQLLK1828 PMCEVKTDFC1838 QTVLPYLIHD 1848 ILLQDTNESW1858 RNLLSTHVQG1868 FFTSCLRHCC1900 LDKKSQRTML1910 AVVDYMRRQK 1920 RPSSGTIFND1930 AFWLDLNYLE1940 VAKVAQSCAA1950 HFTALLYAEI1960 YADKKSMDDQ 1970 EKRSTTISSL1989 SEKSKEETGI1999 SLQDLLLEIY2009 RSIGEPDSLY2019 GCGGGKMLQP 2029 ITRLRTYEHE2039 AMWGKALVTY2049 DLETAIPSST2059 RQAGIIQALQ2069 NLGLCHILSV 2079 YLKGLDYENK2089 DWCPELEELH2099 YQAAWRNMQW2109 DHCGTSYHES2127 LYNALQSLRD 2137 REFSTFYESL2147 KYARVKEVEE2157 MCKRSLESVY2167 SLYPTLSRLQ2177 AIGELESIGE 2187 LFSRSVTHRQ2197 LSEVYIKWQK2207 HSQLLKDSDF2217 SFQEPIMALR2227 TVILEILMEK 2237 EMDNSQRECI2247 KDILTKHLVE2257 LSILARTFKN2267 TQLPERAIFQ2277 IKQYNSVSCG 2287 VSEWQLEEAQ2297 VFWAKKEQSL2307 ALSILKQMIK2317 KLDASCAANN2327 PSLKLTYTEC 2337 LRVCGNWLAE2347 TCLENPAVIM2357 QTYLEKAVEV2367 AGNYDGESSD2377 ELRNGKMKAF 2387 LSLARFSDTQ2397 YQRIENYMKS2407 SEFENKQALL2417 KRAKERYTVK2440 VQRELELDEL 2450 ALRALKEDRK2460 RFLCKAVENY2470 INCLLSGEEH2480 DMWVFRLCSL2490 WLENSGVSEV 2500 NGMMKRDGMK2510 IPTYKFLPLM2520 YQLAARMGTK2530 MMGGLGFHEV2540 LNNLISRISM 2550 DHPHHTLFII2560 LALANANRDE2570 FLTSSQLDED2597 RTEAANRIIC2607 TIRSRRPQMV 2617 RSVEALCDAY2627 IILANLDATQ2637 WKTQRKGINI2647 PADQPITKLK2657 NLEDVVVPTM 2667 EIKVDHTGEY2677 GNLVTIQSFK2687 AEFRLAGGVN2697 LPKIIDCVGS2707 DGKERRQLVK 2717 GRDDLRQDAV2727 MQQVFQMCNT2737 LLQRNTETRK2747 RKLTICTYKV2757 VPLSQRSGVL 2767 EWCTGTVPIG2777 EFLVNNEDGA2787 HKRYRPNDFS2797 AFQCQKKMME2807 VQKKSFEEKY 2817 EVFMDVCQNF2827 QPVFRYFCME2837 KFLDPAIWFE2847 KRLAYTRSVA2857 TSSIVGYILG 2867 LGDRHVQNIL2877 INEQSAELVH2887 IDLGVAFEQG2897 KILPTPETVP2907 FRLTRDIVDG 2917 MGITGVEGVF2927 RRCCEKTMEV2937 MRNSQETLLT2947 IVEVLLYDPL2957 FDWTMNPLKA 2967 LYLQQRPSFN3003 KVAERVLMRL3013 QEKLKGVEEG3023 TVLSVGGQVN3033 LLIQQAIDPK 3043 NLSRLFPGWK3053 AWV
|
|||||
|
|
ALA2693
4.239
GLY2694
3.436
GLY2695
4.218
VAL2696
4.171
ASN2697
4.069
PRO2699
4.858
LEU2715
3.786
LYS2717
3.014
ASP2720
4.489
TYR2755
3.901
LEU2767
3.356
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: KU-55933 | Ligand Info | |||||
| Structure Description | Human ATM kinase with bound inhibitor KU-55933 | PDB:7NI5 | ||||
| Method | Electron microscopy | Resolution | 2.78 Å | Mutation | No | [6] |
| PDB Sequence |
LVLNDLLICC
12 RQLEHDRATE22 RKKEVEKFKR32 LIRDPETIKH42 LDRHSDSKQG52 KYLNWDAVFR 62 FLQKYIQKET72 ECLRIASTQA88 SRQKKMQEIS98 SLVKYFIKCA108 NRRAPRLKCQ 118 ELLNYIMDTV128 KDSSNGAIYG138 ADCSNILLKD148 ILSVRKYWCE158 ISQQQWLELF 168 SVYFRLYLKP178 SQDVHRVLVA188 RIIHAVTKGC198 CSQTDGLNSK208 FLDFFSKAIQ 218 CARQEKSSSG228 LNHILAALTI238 FLKTLAVNFR248 IRVCELGDEI258 LPTLLYIWTQ 268 HRLNDSLKEV278 IIELFQLQIY288 IHHPKGAKTQ298 EKGAYESTKW308 RSILYNLYDL 318 LVNEISHIGS328 RGKYSSGFRN338 IAVKENLIEL348 MADICHQVFI389 ELGWEVIKDH 399 LQKSQNDFDL409 VPWLQIATQL419 ISKYPASLPN429 CELSPLLMIL439 SQLLPQQRHG 449 ERTPYVLRCL459 TEVALCQDKR469 SNLESSQKSD479 LLKLWNKIWC489 ITFRGISSEQ 499 IQAENFGLLG509 AIIQGSLVEV519 DREFWKLFTG529 SACRPSCPAV539 CCLTLALTTS 549 IVPGTFSLKE574 SIMKWLLFYQ584 EVPPILHSNF603 PHLVLEKILV613 SLTMKNCKAA 623 MNFFQSVPEC633 EHHQKDKEEL643 SFSEVEELFL653 QTTFDKMDFL663 TSSIGFSVHQ 684 NLKESLDRCL694 LGLSEQLLNN704 YSSEITNSET714 LVRCSRLLVG724 VLGCYCYMGV 734 IAEEEAYKSE744 LFQKAKSLMQ754 CAGESITLFK764 NKTNEEFRIG774 SLRNMMQLCT 784 RCLSNCTKKS794 PNKIASGFFL804 RLLTSKLMND814 IADICKSLAS824 FIKKIGAINP 884 LAEEYLSKQD894 LLFLDMLKFL904 CLCVTTAQTN914 TVSFRAADIR924 RKLLMLIDSS 934 TLEPTKSLHL944 HMYLMLLKEL954 PGEEYPLPME964 DVLELLKPLS974 NVCSLYRRDQ 984 DVCKTILNHV994 LHVVKNLGQS1004 NMDSENTRDA1014 QGQFLTVIGA1024 FWHLTKERKY 1034 IFSVRMALVN1044 CLKTLLEADP1054 YSKWAILNVM1064 GKDFPVNEVF1074 TQFLADNHHQ 1084 VRMLAAESIN1094 RLFQDTLLKA1110 LPLKLQQTAF1120 ENAYLKAQEG1130 MREMSHSAEN 1140 PETLDEIYNR1150 KSVLLTLIAV1160 VLSCSPICEK1170 QALFALCKSV1180 KENGLEPHLV 1190 KKVLEKVSET1200 FGYRRLEDFM1210 ASHLDYLVLE1220 WLNLQDTEYN1230 LSSFPFILLN 1240 YTNIEDFYRS1250 CYKVLIPHLV1260 IRSHFDEVKS1270 IANQIQEDWK1280 SLLTDCFPKI 1290 LVNILPYFAY1300 EGTRDSGMAQ1310 QRETATKVYD1320 MLKSENLLGK1330 QIDHLFISNL 1340 PEIVVELLMT1350 LHEPDLDPAP1376 NPPHFPSHVI1386 KATFAYISNC1396 ILEILSKSPD 1413 SYQKILLAIC1423 EQAAETNNVY1433 KKHRILKIYH1443 LFVSLLLKDI1453 KSGLGGAWAF 1463 VLRDVIYTLI1473 HYINQRPSCI1483 MDVSLRSFSL1493 CCDLLSQVCQ1503 TAVTYCKDAL 1513 ENHLHVIVGT1523 LIPLVYEQVE1533 VQKQVLDLLK1543 YLVIDNKDNE1553 NLYITIKLLD 1563 PFPDHVVFKD1573 LRITQQKIKY1583 SRGPFSLLEE1593 INHFLSVSVY1603 DALPLTRLEG 1613 LKDLRRQLEL1623 HKDQMVDIMR1633 ASQDNPQDGI1643 MVKLVVNLLQ1653 LSKMAINHTG 1663 EKEVLEAVGS1673 CLGEVGPIDF1683 STIAIQHSKD1693 ASYTKALKLF1703 EDKELQWTFI 1713 MLTYLNNTLV1723 EDCVKVRSAA1733 VTCLKNILAT1743 KTGHSFWEIY1753 KMTTDPMLAY 1763 LQPFRTSRKK1773 FLEVPRFDKE1783 NPFEGLDDIN1793 LWIPLSENHD1803 IWIKTLTCAF 1813 LDSGGTKCEI1823 LQLLKPMCEV1833 KTDFCQTVLP1843 YLIHDILLQD1853 TNESWRNLLS 1863 THVQGFFTSC1873 LRHHFFRCCL1901 DKKSQRTMLA1911 VVDYMRRQKR1921 PSSGTIFNDA 1931 FWLDLNYLEV1941 AKVAQSCAAH1951 FTALLYAEIY1961 ADKKSMDDQE1971 KISSLSEKSK 1994 EETGISLQDL2004 LLEIYRSIGE2014 PDSLYGCGGG2024 KMLQPITRLR2034 TYEHEAMWGK 2044 ALVTYDLETA2054 IPSSTRQAGI2064 IQALQNLGLC2074 HILSVYLKGL2084 DYENKDWCPE 2094 LEELHYQAAW2104 RNMQWDHCGT2122 SYHESLYNAL2132 QSLRDREFST2142 FYESLKYARV 2152 KEVEEMCKRS2162 LESVYSLYPT2172 LSRLQAIGEL2182 ESIGELFSRS2192 VTHRQLSEVY 2202 IKWQKHSQLL2212 KDSDFSFQEP2222 IMALRTVILE2232 ILMEKEMDNS2242 QRECIKDILT 2252 KHLVELSILA2262 RTFKNTQLPE2272 RAIFQIKQYN2282 SVSCGVSEWQ2292 LEEAQVFWAK 2302 KEQSLALSIL2312 KQMIKKLDAS2322 CAANNPSLKL2332 TYTECLRVCG2342 NWLAETCLEN 2352 PAVIMQTYLE2362 KAVEVASDEL2379 RNGKMKAFLS2389 LARFSDTQYQ2399 RIENYMKSSE 2409 FENKQALLKR2419 QRELELDELA2451 LRALKEDRKR2461 FLCKAVENYI2471 NCLLSGEEHD 2481 MWVFRLCSLW2491 LENSGVSEVN2501 GMMKRDGMKI2511 PTYKFLPLMY2521 QLAARMGTKM 2531 MGGLGFHEVL2541 NNLISRISMD2551 HPHHTLFIIL2561 ALANANRDEF2571 LTKSQLDEDR 2598 TEAANRIICT2608 IRSRRPQMVR2618 SVEALCDAYI2628 ILANLDATQW2638 KTQRKGINIP 2648 ADQPITKLKN2658 LEDVVVPTME2668 IKVDHTGEYG2678 NLVTIQSFKA2688 EFRLAGGVNL 2698 PKIIDCVGSD2708 GKERRQLVKG2718 RDDLRQDAVM2728 QQVFQMCNTL2738 LQRNTETRKR 2748 KLTICTYKVV2758 PLSQRSGVLE2768 WCTGTVPIGE2778 FLVNNEDGAH2788 KRYRPNDFSA 2798 FQCQKKMMEV2808 QKKSFEEKYE2818 VFMDVCQNFQ2828 PVFRYFCMEK2838 FLDPAIWFEK 2848 RLAYTRSVAT2858 SSIVGYILGL2868 GDRHVQNILI2878 NEQSAELVHI2888 DLGVAFEQGK 2898 ILPTPETVPF2908 RLTRDIVDGM2918 GITGVEGVFR2928 RCCEKTMEVM2938 RNSQETLLTI 2948 VEVLLYDPLF2958 DWTMNPLKAL2968 YLQQQSFNKV3005 AERVLMRLQE3015 KLKGVEEGTV 3025 LSVGGQVNLL3035 IQQAIDPKNL3045 SRLFPGWKAW3055 V
|
|||||
|
|
ALA2693
3.772
GLY2694
4.223
GLY2695
4.235
PRO2699
3.548
LEU2715
3.906
LYS2717
3.454
ASP2725
4.126
TYR2755
3.824
LEU2767
3.765
GLU2768
3.594
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Homologous recombination | hsa03440 | Affiliated Target |
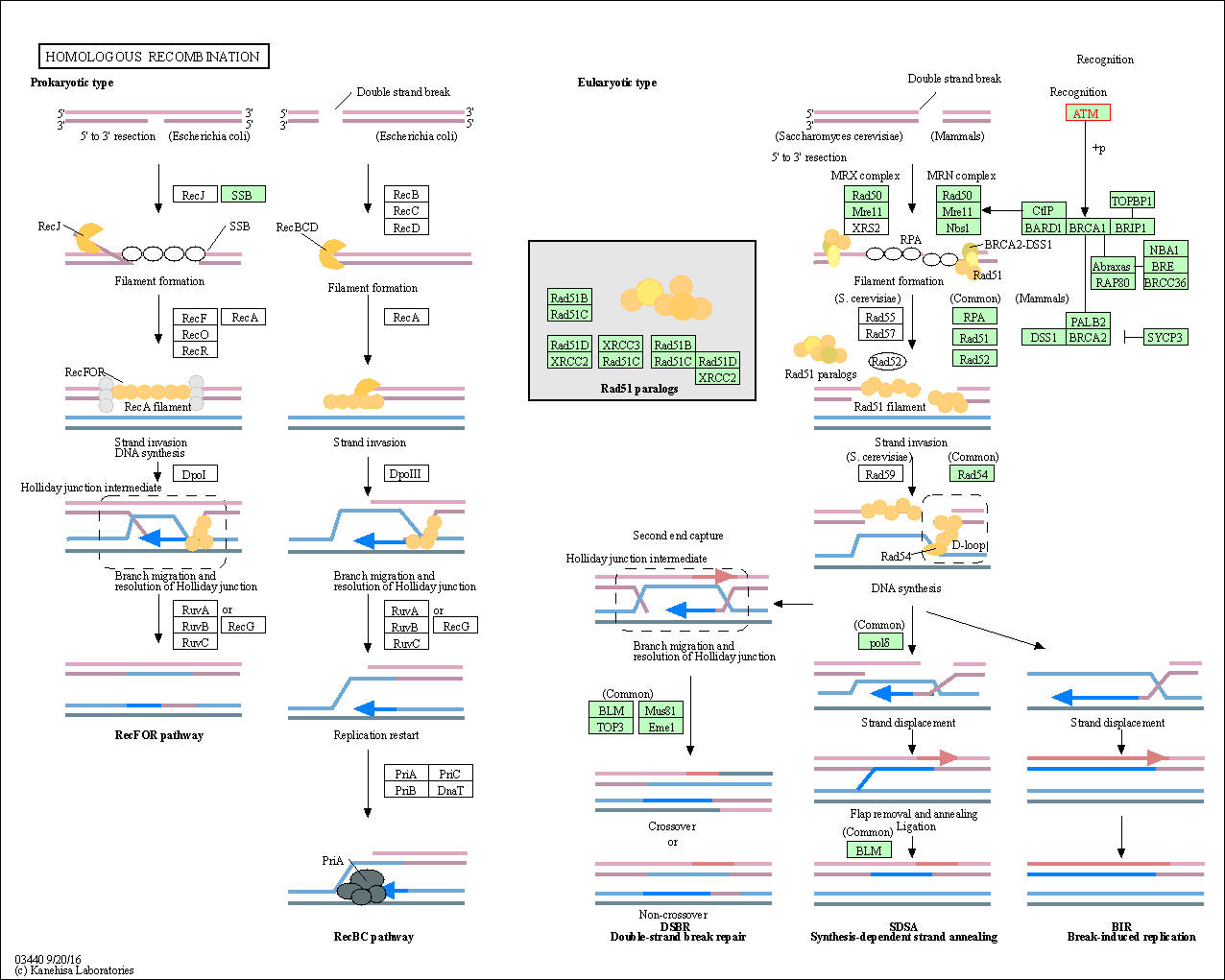
|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
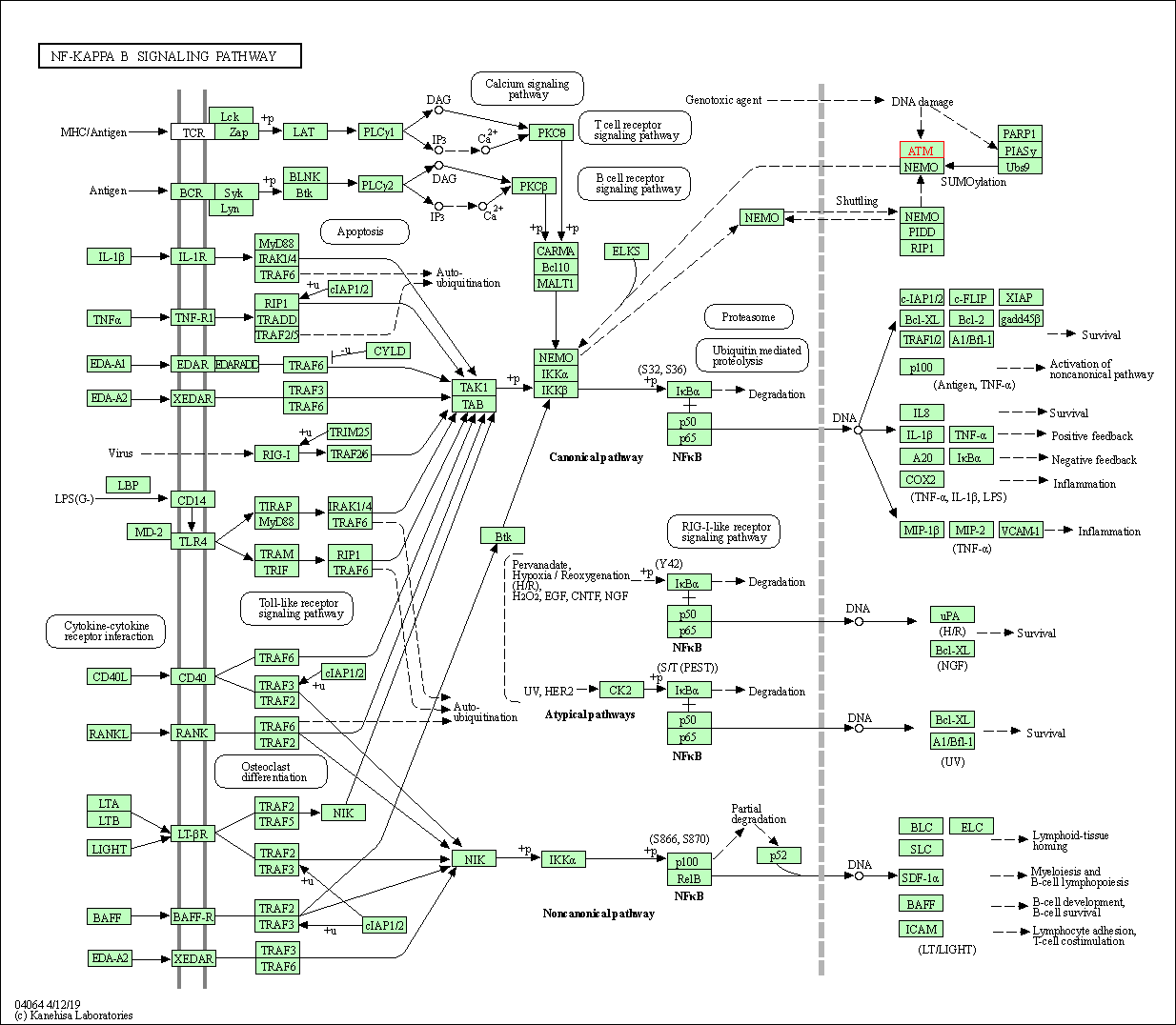
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |
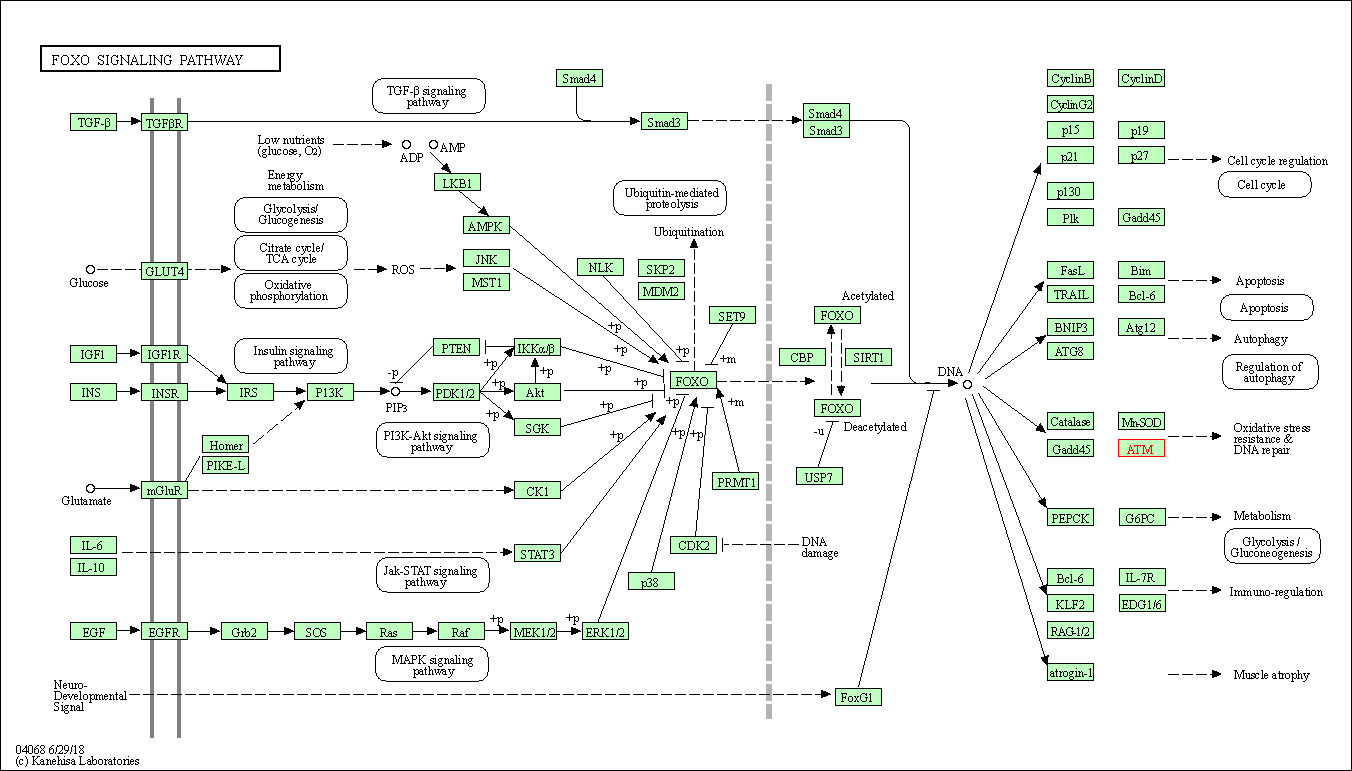
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cell cycle | hsa04110 | Affiliated Target |
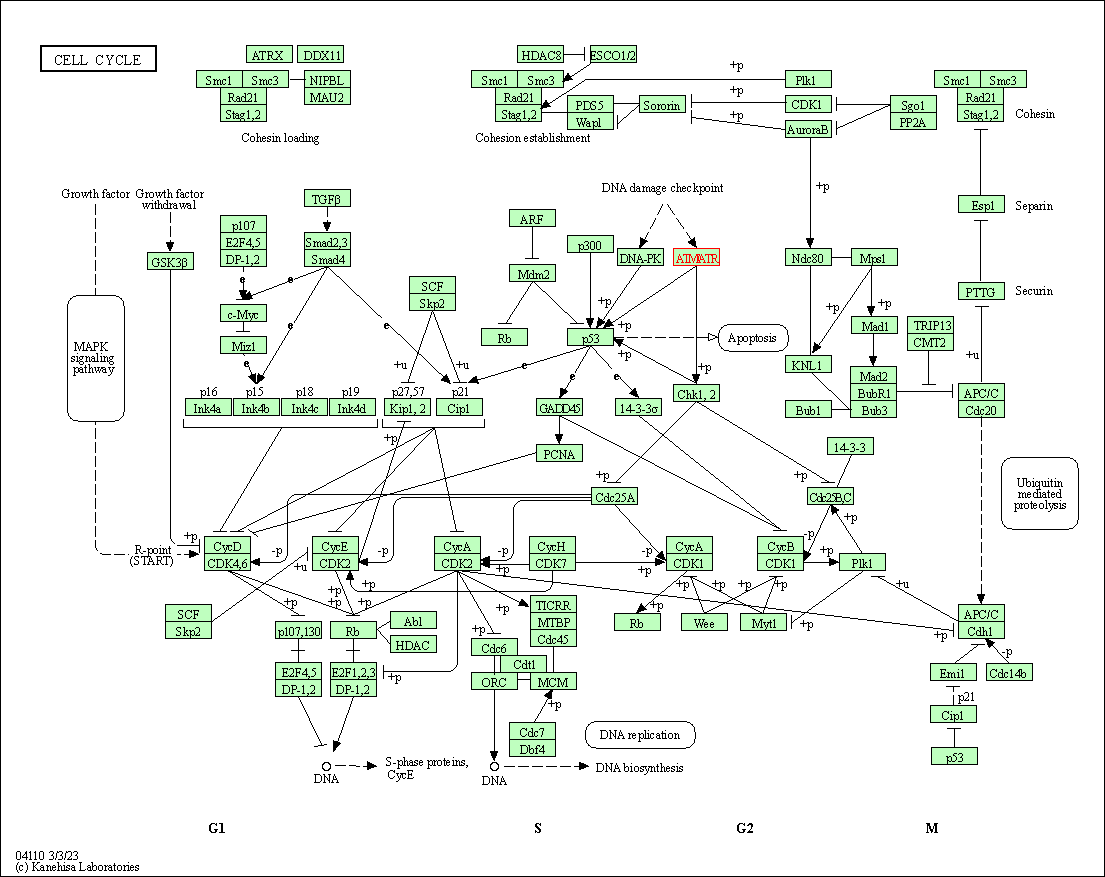
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| p53 signaling pathway | hsa04115 | Affiliated Target |
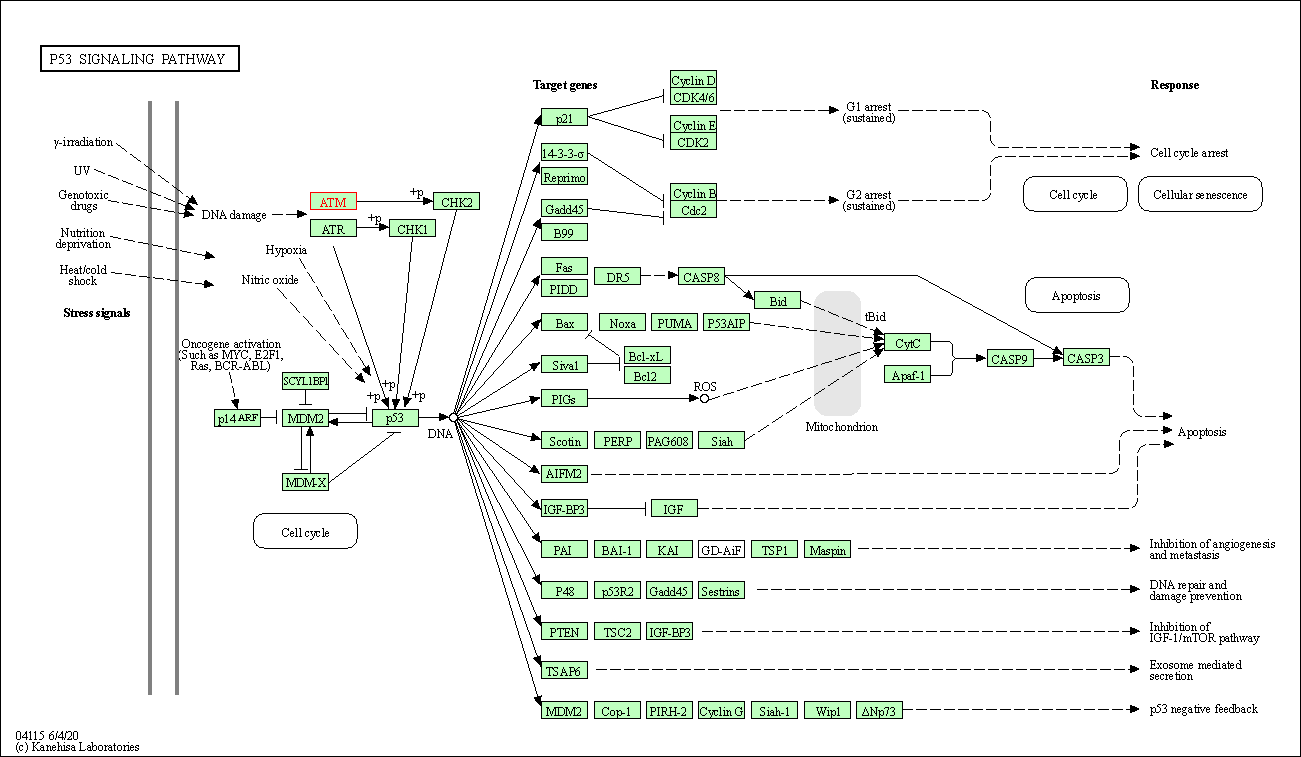
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
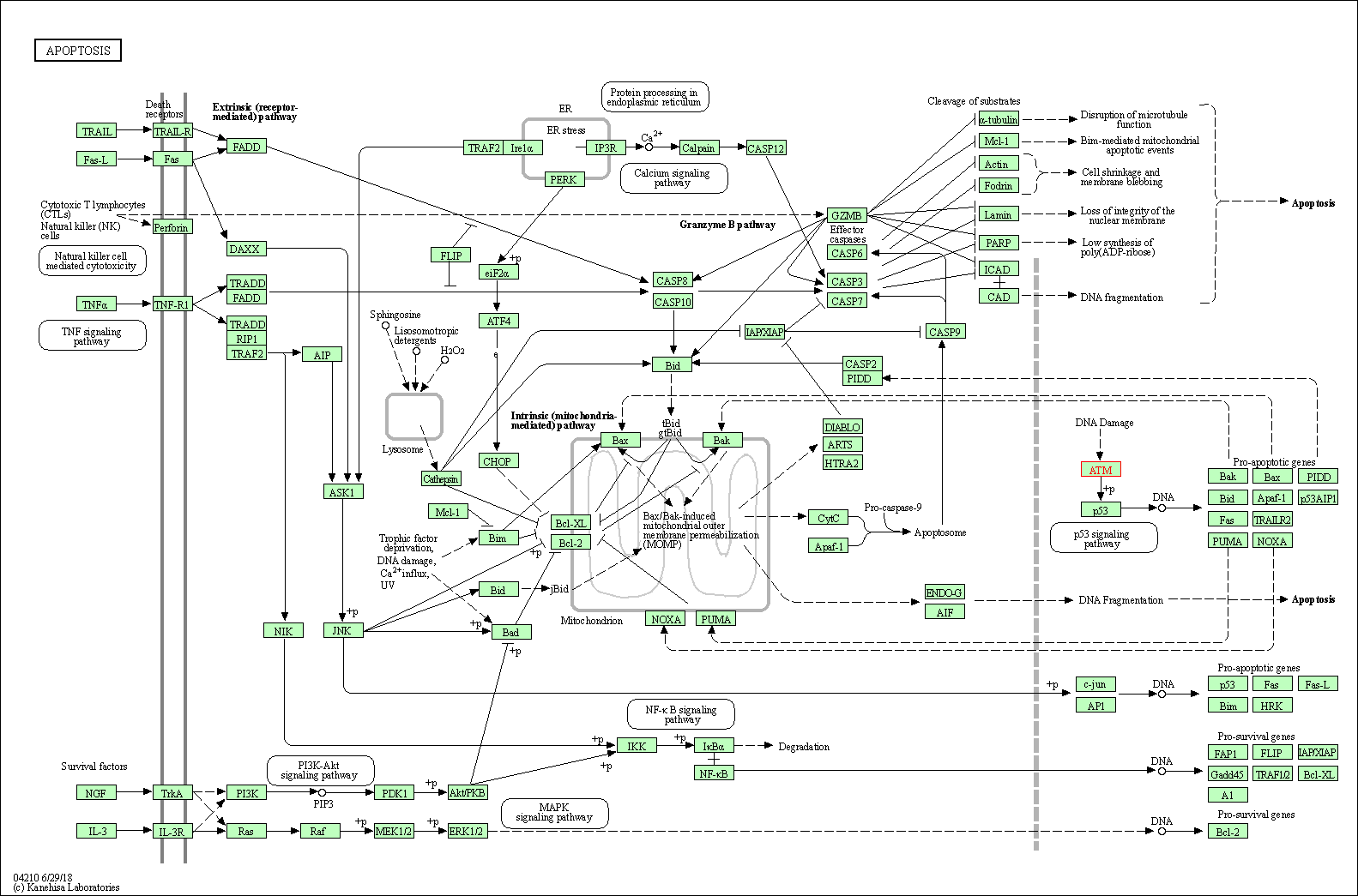
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
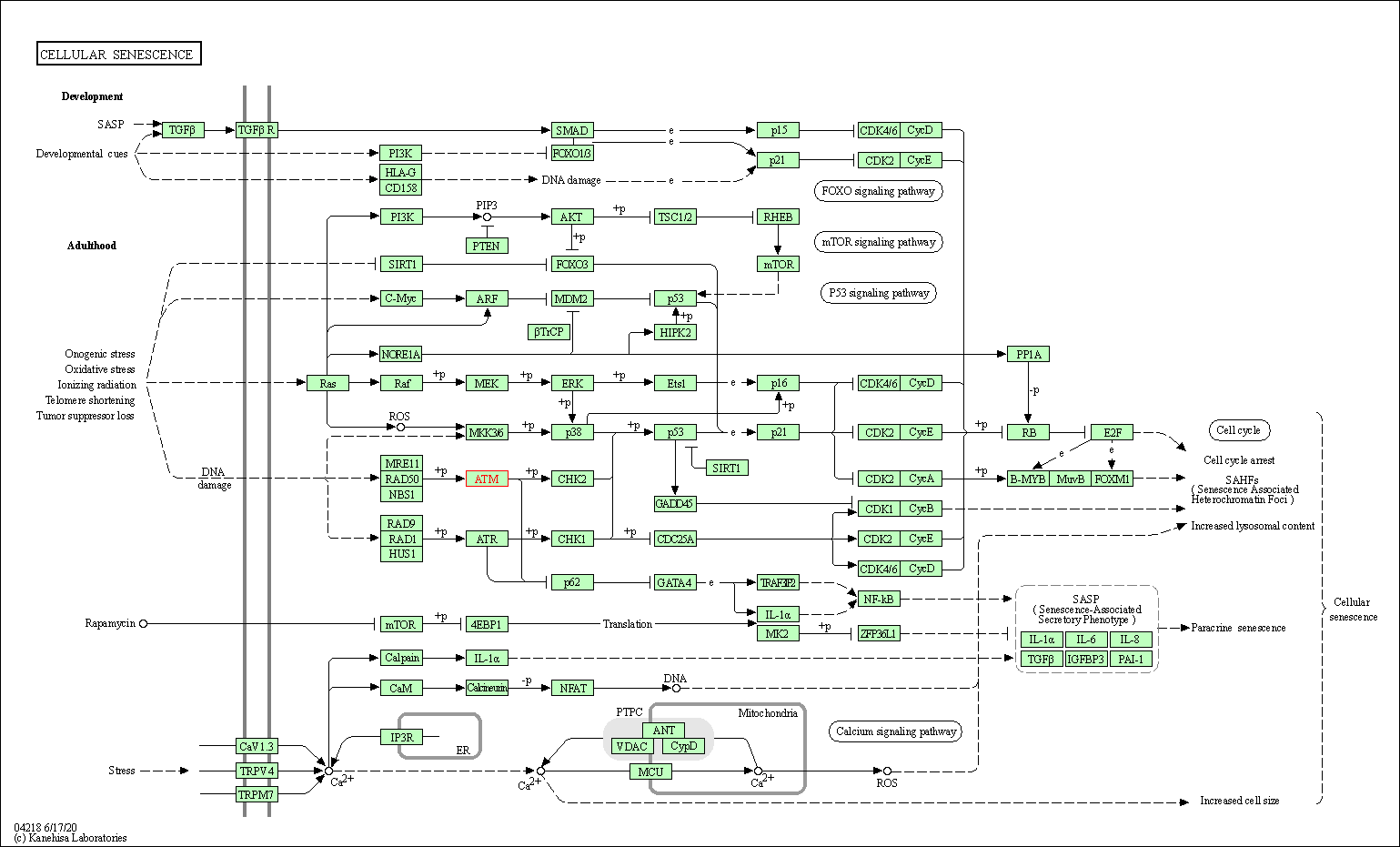
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 79 | Degree centrality | 8.49E-03 | Betweenness centrality | 9.96E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.58E-01 | Radiality | 1.45E+01 | Clustering coefficient | 1.76E-01 |
| Neighborhood connectivity | 3.71E+01 | Topological coefficient | 3.83E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Drug Resistance Mutation (DRM) | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012 Dec 6;3:e441. | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009 Oct;8(10):2894-902. | |||||
| REF 4 | Small molecule-based reversible reprogramming of cellular lifespan. Nat Chem Biol. 2006 Jul;2(7):369-74. | |||||
| REF 5 | Structure of the human ATM kinase and mechanism of Nbs1 binding. Elife. 2022 Jan 25;11:e74218. | |||||
| REF 6 | Molecular basis of human ATM kinase inhibition. Nat Struct Mol Biol. 2021 Oct;28(10):789-798. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

