Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T26203
(Former ID: TTDI01933)
|
|||||
| Target Name |
Intercellular adhesion molecule ICAM-1 (ICAM1)
|
|||||
| Synonyms |
Major group rhinovirus receptor; Intercellular adhesion molecule 1; ICAM-1; CD54
Click to Show/Hide
|
|||||
| Gene Name |
ICAM1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Visual system disease [ICD-11: 9E1Z] | |||||
| Function |
During leukocyte trans-endothelial migration, ICAM1 engagement promotes the assembly of endothelial apical cups through ARHGEF26/SGEF and RHOG activation. ICAM proteins are ligands for the leukocyte adhesion protein LFA-1 (integrin alpha-L/beta-2).
Click to Show/Hide
|
|||||
| BioChemical Class |
Immunoglobulin
|
|||||
| UniProt ID | ||||||
| Sequence |
MAPSSPRPALPALLVLLGALFPGPGNAQTSVSPSKVILPRGGSVLVTCSTSCDQPKLLGI
ETPLPKKELLLPGNNRKVYELSNVQEDSQPMCYSNCPDGQSTAKTFLTVYWTPERVELAP LPSWQPVGKNLTLRCQVEGGAPRANLTVVLLRGEKELKREPAVGEPAEVTTTVLVRRDHH GANFSCRTELDLRPQGLELFENTSAPYQLQTFVLPATPPQLVSPRVLEVDTQGTVVCSLD GLFPVSEAQVHLALGDQRLNPTVTYGNDSFSAKASVSVTAEDEGTQRLTCAVILGNQSQE TLQTVTIYSFPAPNVILTKPEVSEGTEVTVKCEAHPRAKVTLNGVPAQPLGPRAQLLLKA TPEDNGRSFSCSATLEVAGQLIHKNQTRELRVLYGPRLDERDCPGNWTWPENSQQTPMCQ AWGNPLPELKCLKDGTFPLPIGESVTVTRDLEGTYLCRARSTQGEVTRKVTVNVLSPRYE IVIITVVAAAVIMGTAGLSTYLYNRQRKIKKYRLQQAQKGTPMKPNTQATPP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A02322 ; BADD_A02324 ; BADD_A05290 ; BADD_A05438 ; BADD_A06206 ; BADD_A06414 ; BADD_A08478 | |||||
| HIT2.0 ID | T10HL8 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | lifitegrast | Drug Info | Approved | Dry eye disease | [2] | |
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | Alicaforsen | Drug Info | Phase 3 | Lleum inflammation | [3] | |
| 2 | BI-505 | Drug Info | Phase 2 | Multiple myeloma | [3] | |
| Discontinued Drug(s) | [+] 3 Discontinued Drugs | + | ||||
| 1 | A-252444.0 | Drug Info | Terminated | Inflammation | [6] | |
| 2 | GI-270384X | Drug Info | Terminated | Inflammatory bowel disease | [7] | |
| 3 | MOR-102 | Drug Info | Terminated | Psoriasis vulgaris | [8] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | lifitegrast | Drug Info | [1] | |||
| Antagonist | [+] 2 Antagonist drugs | + | ||||
| 1 | Alicaforsen | Drug Info | [9] | |||
| 2 | A-252444.0 | Drug Info | [11] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | GI-270384X | Drug Info | [7] | |||
| 2 | ISIS-1939 | Drug Info | [13] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Myo-inositol hexaphosphate | Ligand Info | |||||
| Structure Description | The DBLb domain of PF11_0521 PfEMP1 bound to human ICAM-1 | PDB:5MZA | ||||
| Method | X-ray diffraction | Resolution | 2.78 Å | Mutation | No | [14] |
| PDB Sequence |
QTSVSPSKVI
10 LPRGGSVLVT20 CSTSCDQPKL30 LGIETPLPKK40 ELLLPGNNRK50 VYELSNVQED 60 SQPMCYSNCP70 DGQSTAKTFL80 TVYWTPERVE90 LAPLPSWQPV100 GKNLTLRCQV 110 EGGAPRANLT120 VVLLRGEKEL130 KREPAVGEPA140 EVTTTVLVRR150 DHHGANFSCR 160 TELDLRPQGL170 ELFENTSAPY180 QLQTFG
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
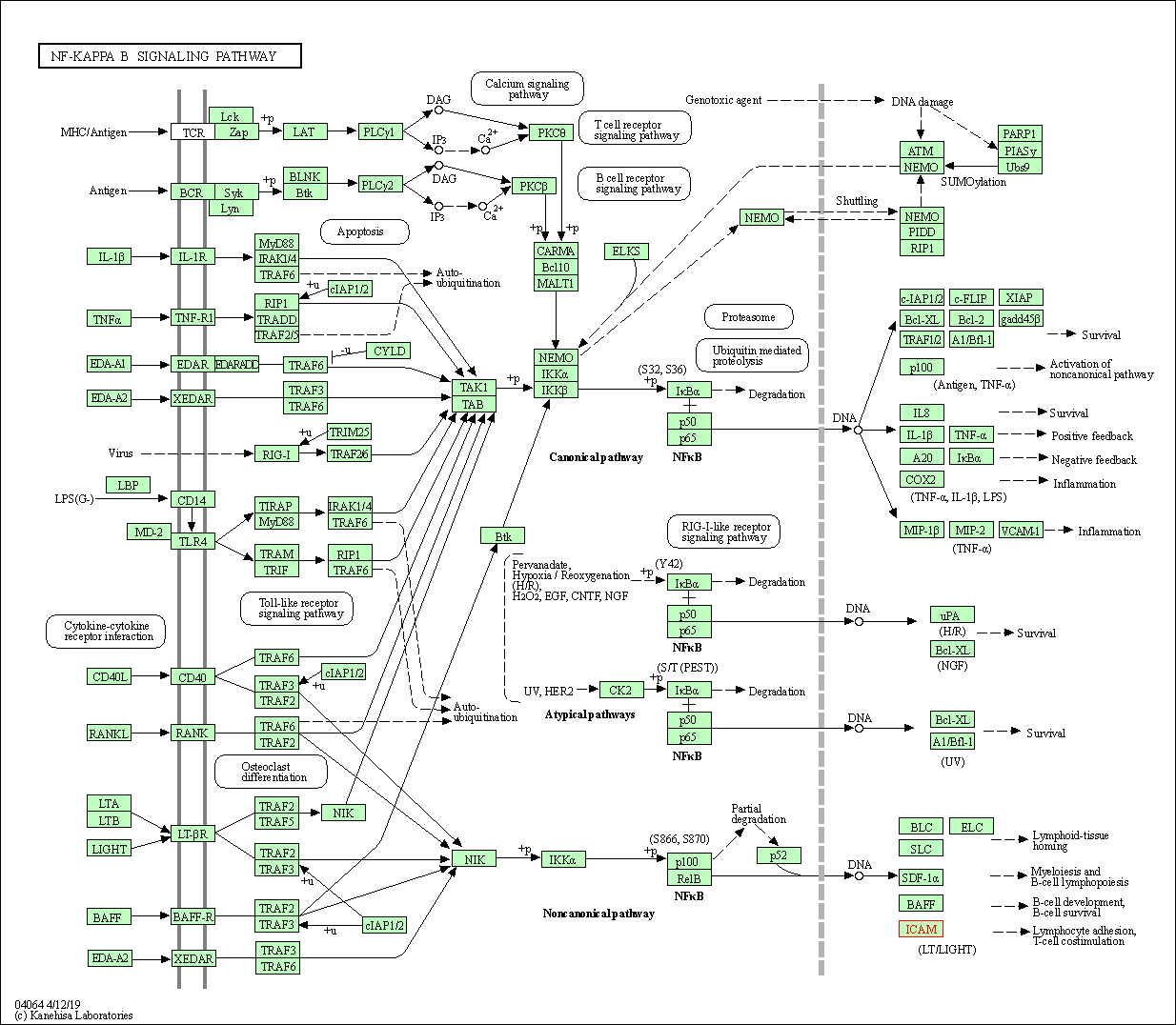
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cell adhesion molecules | hsa04514 | Affiliated Target |
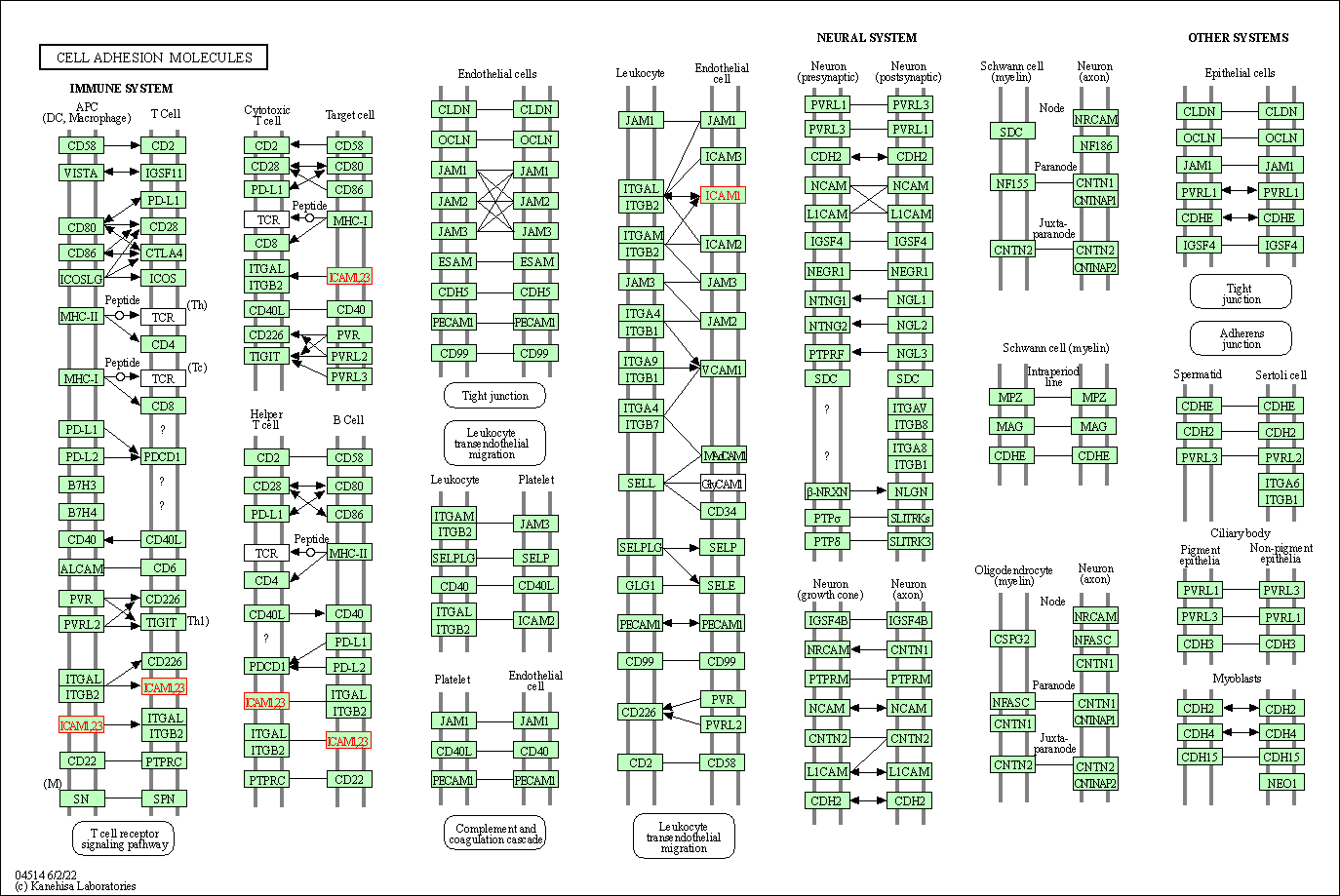
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |
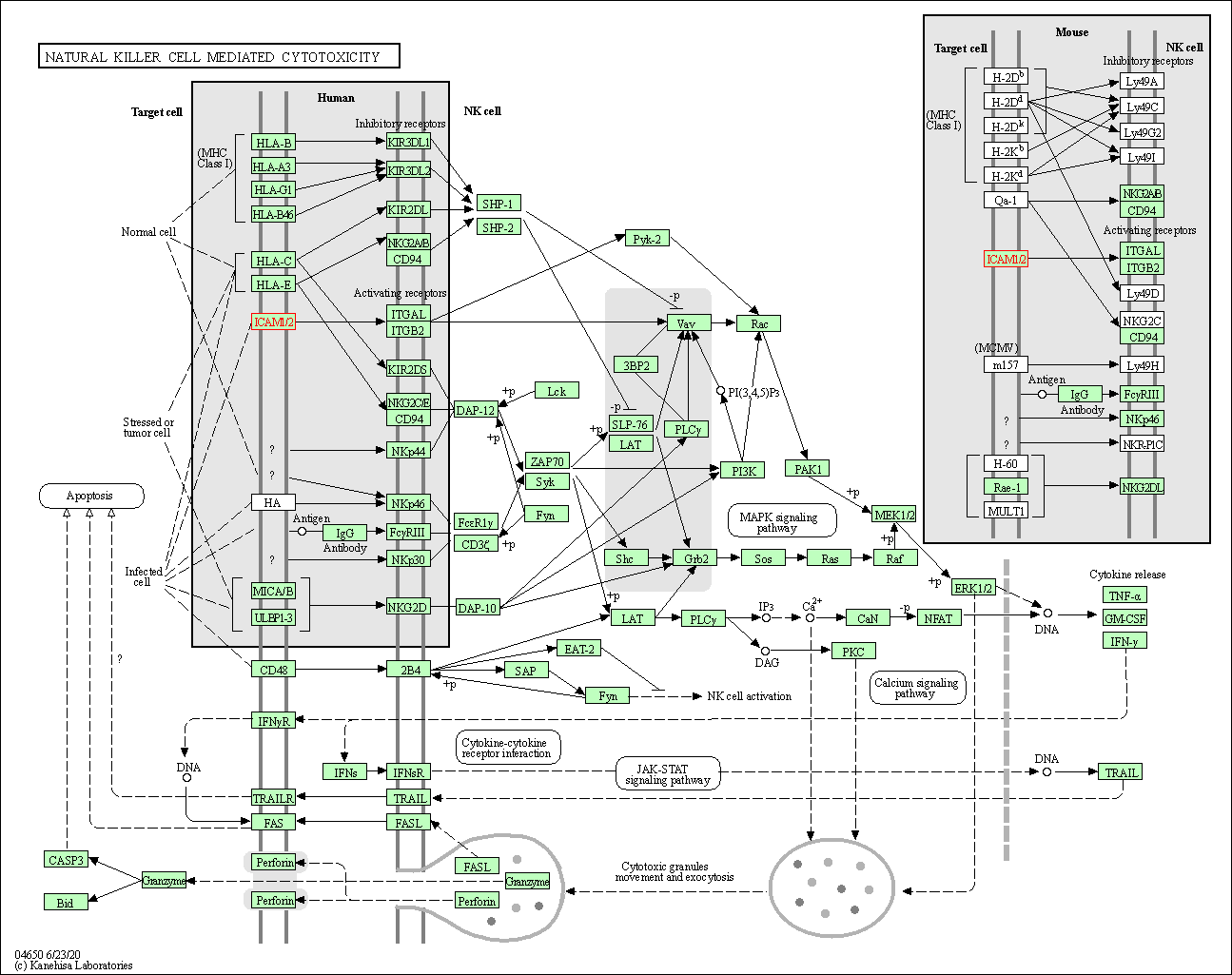
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
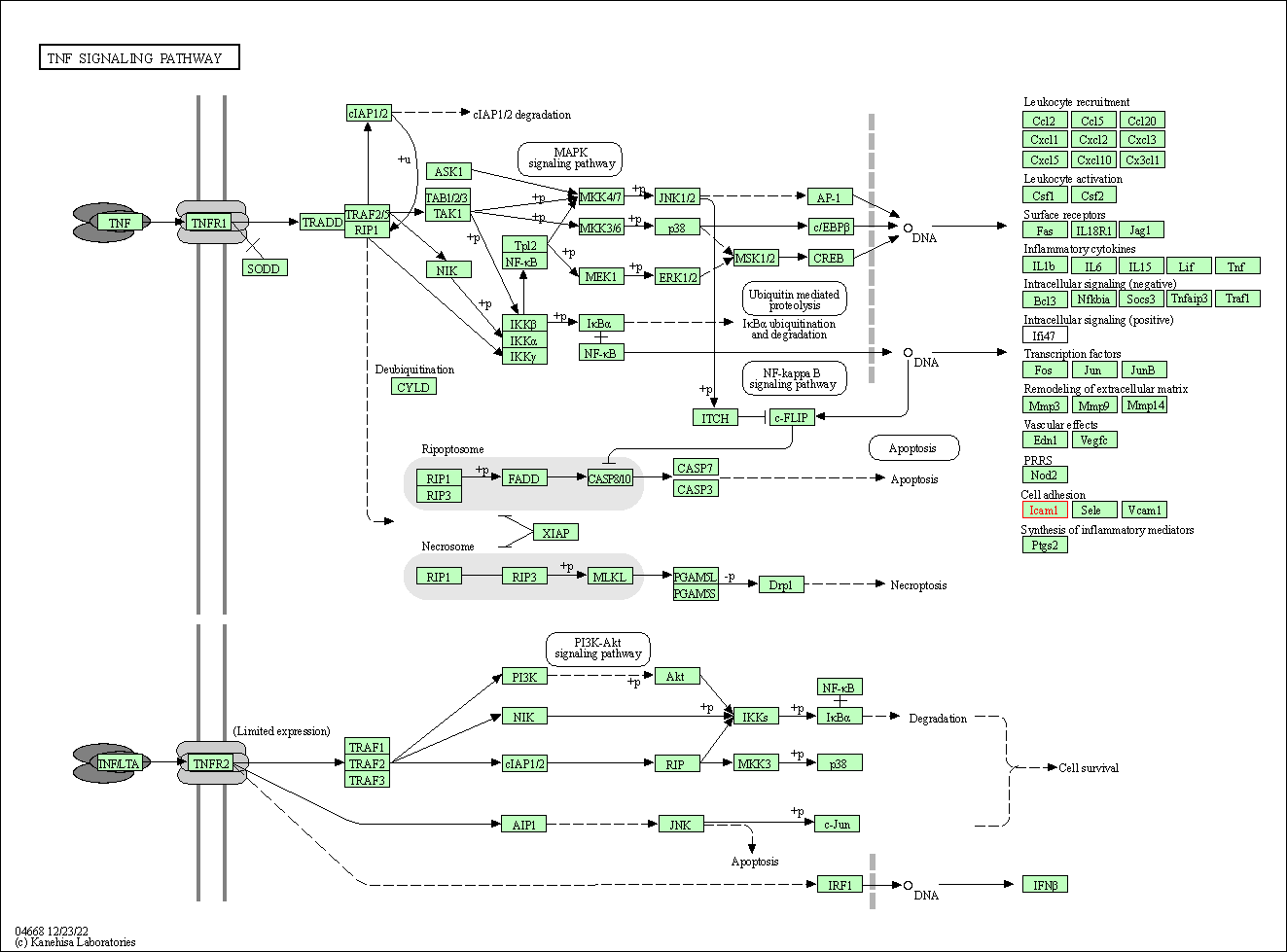
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Leukocyte transendothelial migration | hsa04670 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 16 | Degree centrality | 1.72E-03 | Betweenness centrality | 6.37E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.28E-01 | Radiality | 1.40E+01 | Clustering coefficient | 1.75E-01 |
| Neighborhood connectivity | 2.81E+01 | Topological coefficient | 8.90E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Discovery and Development of Potent LFA-1/ICAM-1 Antagonist SAR 1118 as an Ophthalmic Solution for Treating Dry Eye. ACS Med Chem Lett. 2012 Jan 31;3(3):203-6. | |||||
| REF 2 | 2016 FDA drug approvals. Nat Rev Drug Discov. 2017 Feb 2;16(2):73-76. | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | ClinicalTrials.gov (NCT00849290) Immunotherapy For Men With Objective Disease Progression On Protocol D9902 Part B (NCT00065442). U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT04420754) Study of AIC100 in Relapsed/Refractory Thyroid Cancer. U.S. National Institutes of Health. | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800012566) | |||||
| REF 7 | Inhibition of endothelial cell adhesion molecule expression improves colonic hyperalgaesia. Neurogastroenterol Motil. 2009 Feb;21(2):189-96. | |||||
| REF 8 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800017213) | |||||
| REF 9 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 10 | A human ICAM-1 antibody isolated by a function-first approach has potent macrophage-dependent antimyeloma activity in vivo. Cancer Cell. 2013 Apr 15;23(4):502-15. | |||||
| REF 11 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800012566) | |||||
| REF 12 | Efficacy of the fully human monoclonal antibody MOR102 (#5) against intercellular adhesion molecule 1 in the psoriasis-severe combined immunodeficient mouse model. Br J Dermatol. 2005 Oct;153(4):758-66. | |||||
| REF 13 | Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J Biol Chem. 1991 Sep 25;266(27):18162-71. | |||||
| REF 14 | Structure-Guided Identification of a Family of Dual Receptor-Binding PfEMP1 that Is Associated with Cerebral Malaria. Cell Host Microbe. 2017 Mar 8;21(3):403-414. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

